Peptidomics-Based Disease Mechanism Research Service
- Non-enzymatic direct analysis to avoid degradation bias
- Support for diverse sample types, including tissues, fluids, microorganisms, and cells
- Enhanced signal detection for low-abundance peptides
- Comprehensive analysis of peptide physicochemical properties
- Flexible data mining and expanded analysis services for target discovery
- Type 1 Diabetes: Identifying autoantibody-related peptides to analyze immune dysregulation mechanisms.
- Type 2 Diabetes: Discovering metabolic regulatory peptides to assist in therapeutic target screening.
Peptidomics is a specialized field focused on studying endogenous peptides and their expression changes in pathological conditions. These peptides, typically with a molecular weight under 10 kDa, include disease-specific peptides, protein degradation products, and functional fragments involved in key biological processes such as signal transduction, immune response, and apoptosis. These peptides play crucial roles in various diseases and offer insights into their mechanisms.
Unlike proteomics, peptidomics provides a more sensitive approach for detecting protein degradation and subtle molecular changes. It is particularly well-suited for understanding the molecular mechanisms behind disease progression, treatment responses, and therapeutic effects. High-throughput quantitative analysis of endogenous peptides using mass spectrometry allows us to identify disease-specific expression patterns, uncover potential biomarkers, and discover drug targets.
MtoZ Biolabs offers a specialized Peptidomics-Based Disease Mechanism Research Service that leverages advanced mass spectrometry platforms, enzyme-free detection methods, high-resolution data acquisition, and comprehensive bioinformatics analysis. We help researchers comprehensively characterize dynamic endogenous peptides associated with disease states, supporting studies on disease mechanisms, biomarker discovery, and precision medicine.
Services at MtoZ Biolabs
MtoZ Biolabs offers customized peptidomics services designed to help clients achieve breakthroughs in disease mechanism research. Our services include:
· High-Throughput Identification and Quantification of Endogenous Peptides
We use high-resolution mass spectrometry for sequence analysis, complemented by circular dichroism (CD) spectroscopy to assess peptide secondary structures. For quantitative analysis, we apply advanced technologies like TMT, SILAC, and Label-Free to accurately measure peptide expression levels, identifying disease-specific molecular characteristics.
· Identification and Functional Analysis of Post-Translational Modification (PTM) Peptides
Using advanced mass spectrometry techniques, we identify post-translational modifications (such as phosphorylation and acetylation) in peptides and explore their roles in disease mechanisms.
· Analysis of Unknown Peptides
For unknown peptides or samples from non-model species, we apply a combination of de novo sequencing and database search strategies to improve identification accuracy.
· Comprehensive Bioinformatics Analysis and Data Interpretation
We analyze peptidomics data using bioinformatics tools, providing insights into peptide composition, isoelectric points, hydrophobicity, and other physicochemical properties to better understand their biological functions.
· Functional Prediction and Molecular Docking Analysis (Optional Service)
Based on client needs, we offer functional predictions for peptides, such as anti-inflammatory, antioxidant, and antimicrobial properties, as well as molecular docking simulations to help researchers identify potential therapeutic targets.
Analysis Workflow
MtoZ Biolabs has developed a standardized, traceable peptidomics workflow that covers each crucial step from sample extraction to data identification, ensuring reproducibility and data reliability.
1. Peptide Extraction
We select the appropriate lysis system based on sample type (e.g., tissue, body fluids, cells, microorganisms), using denaturants, heating, and sonication to release endogenous peptides while minimizing degradation.
2. Sample Pretreatment
We employ solid-phase extraction (SPE), ultrafiltration, or ethanol precipitation to remove macromolecular impurities and enrich target peptides, enhancing sensitivity and quantitative accuracy in mass spectrometry detection.
3. Peptide Purification
Using techniques such as ion exchange, reversed-phase chromatography, or capillary electrophoresis, we further separate and enrich peptides, boosting the detection of low-abundance disease-related peptides.
4. Mass Spectrometry Detection
We use high-resolution mass spectrometry systems, including Orbitrap, MALDI-TOF, and LC-MS/MS, to detect peptides with high sensitivity, capturing multidimensional signal characteristics like mass, charge, and retention time.
5. Peptide Identification
We use database matching and de novo sequencing technologies to determine peptide sequences, supported by CD spectroscopy to assess secondary structures, providing the data foundation for further disease mechanism analysis.
6. Bioinformatics Analysis
We perform quantitative comparisons, differential screening, and functional enrichment analysis of identified peptides, using GO, KEGG pathways, protein interaction networks, and organ-specific expression databases to interpret the biological roles of peptides in disease states, helping with target discovery and mechanism elucidation.
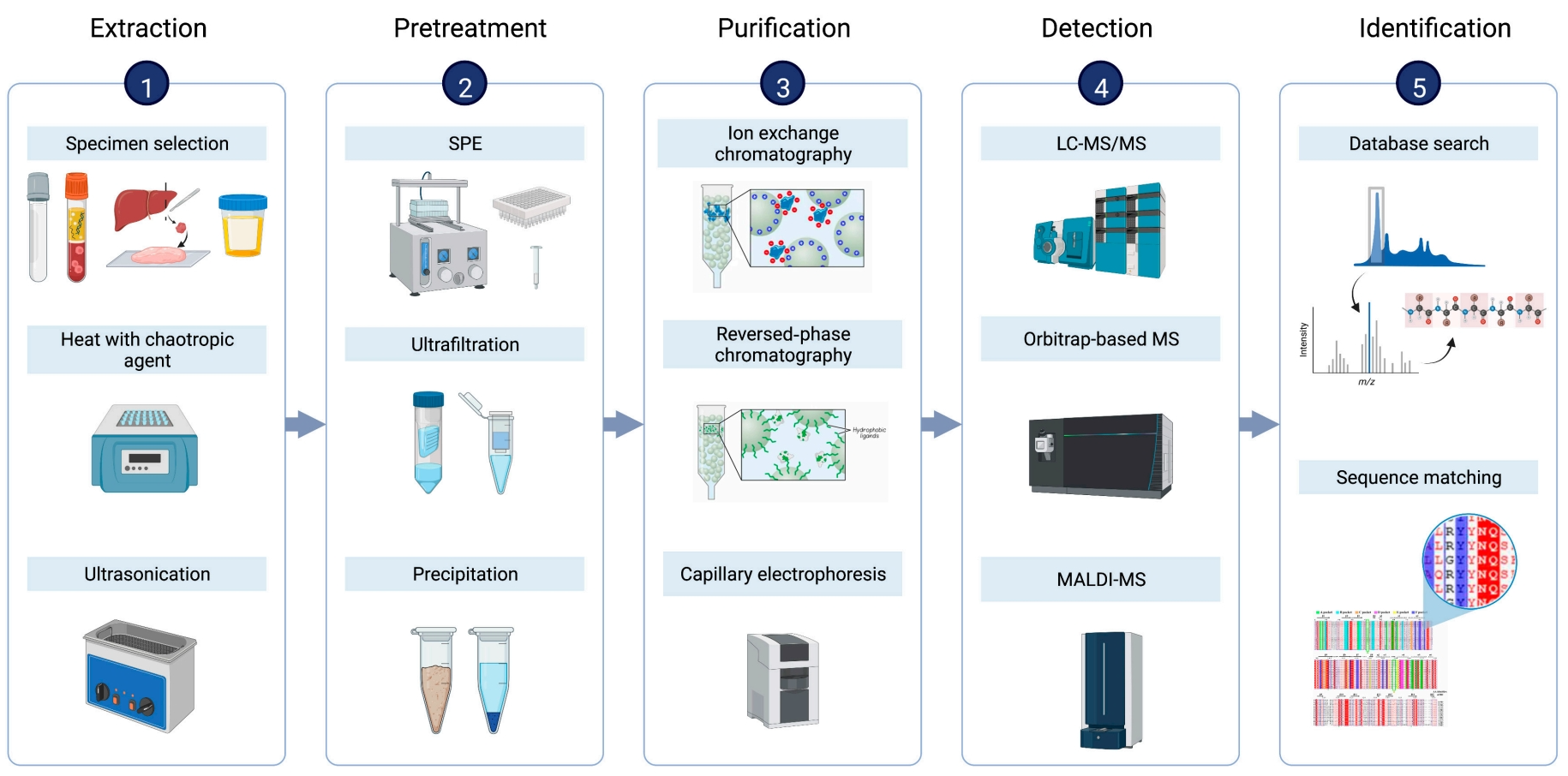
Li, L. et al. BME Front. 2023.
Figure 1. Schematic of Key Steps in Standard Peptidomic Workflows
Why Choose MtoZ Biolabs?
Sample Submission Suggestions
1. Sample Types
Serum, plasma, urine, tissue extracts, and other biological samples.
2. Sample Volume
At least 100 μL of liquid sample or 100 mg of solid sample.
3. Sample Preservation
Store samples at -20°C or lower to prevent degradation, avoiding repeated freeze-thaw cycles.
Note: Provide details on sample collection and handling. If you need further details, our technical support team is happy to assist and provide comprehensive guidance on sample submission.
Applications
Peptidomics-Based Disease Mechanism Research Service is applicable to the following research areas:
· Cancer Research: Identifying cancer-related peptides for early diagnosis, immune monitoring, and efficacy evaluation. Applicable to breast cancer, lung cancer, and other cancer types.
· Diabetes Mechanism Exploration
· Autoimmune Disease Mechanism Research: Uncovering antigenic peptides and immune signaling pathways in diseases such as rheumatoid arthritis and lupus.
· Neurodegenerative Disease Biomarker Discovery: Identifying brain-derived peptides in Alzheimer's and Parkinson's diseases, supporting early diagnosis and intervention research.
· Functional and Antimicrobial Peptide Discovery: Analyzing natural bioactive peptides such as antimicrobial peptides and cytokine fragments, with broad applications in infection control and functional food development.
MtoZ Biolabs is committed to helping global researchers understand the molecular nature of diseases through our high-quality Peptidomics-Based Disease Mechanism Research Service. Our service integrates cutting-edge mass spectrometry platforms with structural recognition and mechanism modeling to offer professional, efficient, and translatable peptidomics research solutions. If you're interested, contact us for a free technical consultation and customized project proposal.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data File
Related Services
Peptidomics-Based Research Service
Peptidomics-Based PROTAC Research Service
Peptidomics-Based Proteolytic Pathways Research Service
Peptidomics-Based Cosmetic Research Service
How to order?







