mRNA Drug Analysis Service
mRNA medication is a novel therapeutic method that utilizes messenger RNA (mRNA). mRNA is a sequence of ribotide that encodes for proteins and can express specific proteins through the translation process within cells. The core principle of mRNA medications is to introduce designed mRNA molecules into the patient's body to produce proteins needed for treatment by utilizing the host cell's translation mechanism, achieving the purpose of treating diseases.
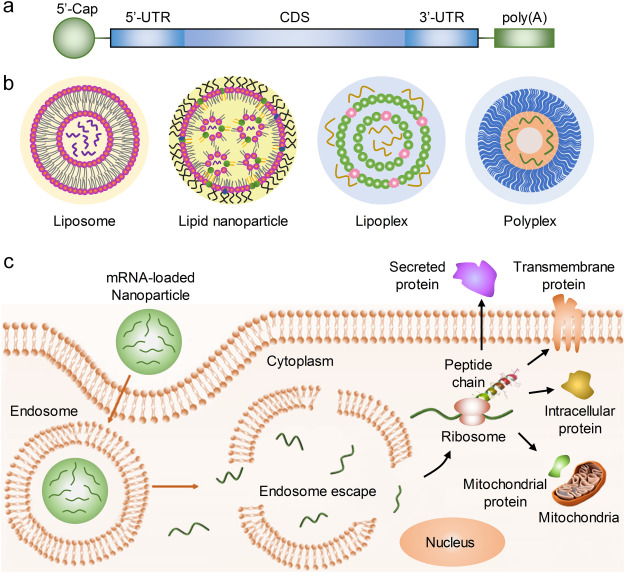
Figure 1. mRNA Delivery and Protein Expression [1]
The development and application of mRNA medication are very broad, mainly including:
1. Infectious Disease Vaccines
mRNA technology has made significant progress in vaccines against infectious diseases, especially during the COVID-19 pandemic. The rapid development and wide usage of mRNA vaccines (such as BNT162b2 and mRNA-1273) have demonstrated the potential and advantage of mRNA technology in vaccine development.
2. Cancer Vaccines
mRNA cancer vaccines activate the immune system against tumor cells by encoding tumor-specific antigens. These vaccines are well-tolerated and safe. They generally cause mild to moderate adverse events.
3. Protein Replacement Therapy
mRNA technology can also be used for protein replacement therapy. For example, replacing missing or defective proteins in metabolic diseases, neurogenic diseases, muscular atrophy and other related diseases to achieve therapeutic goals.
4. Gene Editing
mRNA is also used to deliver programmable nucleases, such as the CRISPR/Cas system, for gene editing treatments, which is characterized by transient expression and no mutated risks.
Advantages and Limitations of mRNA Medications as Therapeutic Drugs
1. Advantages
(1) Rapid development: mRNA medications can be quickly designed and synthesized, especially for vaccine development, as demonstrated during the COVID-19 pandemic.
(2) High specificity: mRNA medications can precisely encode specific proteins for treating specific diseases.
(3) No genomic integration risk: mRNA does not integrate into the host genome, thus avoiding the risk of insertion mutations.
(4) Easy production: The production process of mRNA is relatively simple, cost-effective, and can be scaled up.
(5) Dual immune activation: mRNA vaccines can activate both humoral and cellular immunity.
(6) Therapeutic diversity: mRNA technology can be used not only for developing vaccine but also for treating a variety of diseases such as protein replacement, gene editing, and cancer treatment.
2. Limitations
(1) Delivery challenges: The delivery of mRNA is a major barrier in the development of mRNA medications, as mRNA medications are large molecules and negatively charged, making it difficult to pass through biological membranes.
(2) Stability challenges: mRNA molecules are susceptible to nucleases in the body, requiring special formulation systems to protect the mRNA from degradation.
(3) Immunogenicity challenges: mRNA medications might induce an immune response in patients, although this risk can be reduced through chemical modifications.
(4) Off-target effects: RNA medications might affect the expression of non-target genes, requiring more precise sequence design to minimize this effect.
(5) Production costs: Although the production of mRNA is relatively simple, its high-quality control and purification processes can increase production costs.
(6) Regulatory and approval challenges: As an emerging technology, the regulatory and approval of mRNA medications might face additional challenges and uncertainties.
Strategies to Overcome Limitations Associated with mRNA Medications
1. Chemical Modifications
Chemical modification of exogenous mRNA, such as capping, tailing, using unnatural nucleotides to replace natural ones, etc., can increase the stability of mRNA and reduce its immunogenicity.
2. Delivery System Improvements
Delivery systems like lipid nanoparticles (LNPs) can effectively transport mRNA into cells. Improving the formulation of LNPs, such as adjusting the composition and ratio of lipid, can enhance cell specificity and reduce immunogenicity.
3. Targeted Delivery
Developing delivery systems with tissue specificity, such as changing the lipid components of LNPs or using specific ligands for active targeting, can improve the expression of mRNA medications in specific organs or cells.
4. Circular mRNA (circRNA)
Using circRNA can prevent mRNA degradation by exonucleases, extending the half-life of mRNA within cells, increasing the total protein expression, and avoiding the costly addition of a 5' cap and Poly(A) tail.
5. Self-Amplifying mRNA (saRNA)
Utilizing the self-replicating capabilities of RNA viruses, the dosage and frequency of administration can be reduced, and the amount of mRNA needed can be significantly decreased compared to linear mRNA.
6. Increasing Translation Efficiency
By optimizing codons and functionally regulating the 5' and 3' untranslated regions, the translation efficiency of mRNA can be increased.
7. Reducing double-stranded RNA (dsRNA) Impurities
Removing dsRNA impurities during the mRNA purification process can reduce the immune response of and improve the stability of mRNA.
8. New Delivery Systems
In addition to LNPs, researchers are exploring other delivery systems, such as polymers, polymer-based nanoparticles and improved peptide carriers, to enhance the delivery efficiency and specificity of mRNA medications.
9. Optimizing Preclinical Models
Using more accurate preclinical models to assess delivery effects in non-human primates to improve the clinical success rate.
10. DNA Barcode Systems
Using DNA barcode systems to simultaneously track multiple nanoparticles in animal bodies, improving screening efficiency.
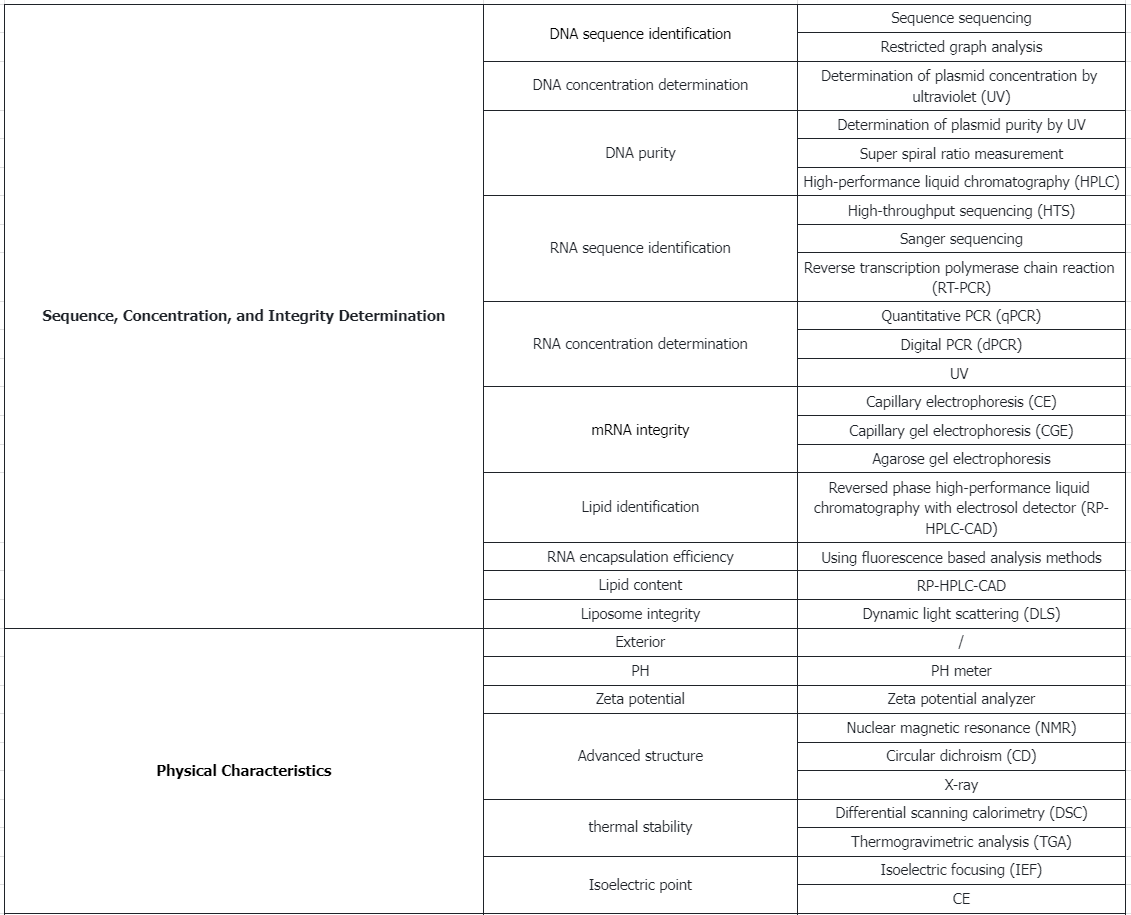
Testing Content Required by Policies and Regulations
Analytical Procedures for mRNA Vaccine Quality 2023
1. Quality Control Metrics and Methods for DNA Plasmids Before Release
(1) Identity
① Method: Sequence sequencing.
② Method: Restriction enzyme analysis with agarose gel electrophoresis.
(2) Concentration
Method: Plasmid concentration (A260) measured by ultraviolet spectroscopy (UV).
(3) Purity
① Method: Plasmid purity (A260/280) measured by ultraviolet spectroscopy (UV).
② Method: Percentage of supercoiled determined by capillary electrophoresis (CE).
③ Method: High-performance liquid chromatography (HPLC).
(4) Residual Host RNA
① Method: HPLC.
② Method: Agarose gel electrophoresis.
(5) Residual Host DNA
Method: Quantitative PCR (qPCR).
(6) Residual Protein
① Method: SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
② Method: Bicinchoninic acid assay (BCA).
(7) Host Cell Protein
Method: Enzyme-linked immunosorbent assay (ELISA).
(8) Residual Kanamycin
Method: ELISA.
(9) Safety
① Method: Endotoxins (USP <85>).
② Method: Bioburden (USP <61>, <62>, <1115>).
③ Method: Sterility (USP <71>).
(10) Other
① Method: Appearance (USP <1>, <790>).
② Method: pH (USP <791>).
Note: USP<number>refers to specific chapters or guidelines in the United States Pharmacopoeia, which provide more detailed standards and testing methods.
2. Characterization and Release Testing of mRNA Drug Substance
(1) Identity
mRNA sequence identity confirmed by high throughput sequencing (HTS), sanger sequencing, or reverse transcriptase polymerase vhain reaction (RT-PCR).
(2) Content
RNA concentration measured by qPCR, digital PCR (dPCR), or UV.
(3) Integrity
mRNA integrity assessed by CE, capillary gel electrophoresis (CGE), or agarose gel electrophoresis.
(4) Purity
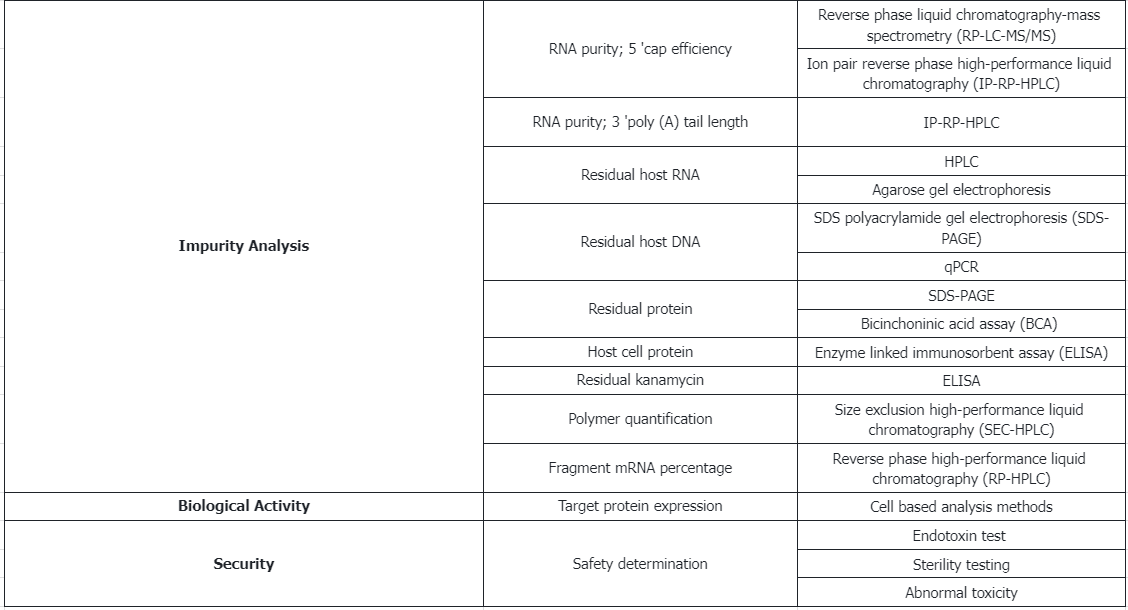
① 5' Cap efficiency determined by reverse-phase liquid chromatography-mass spectrometry (RP–LC-MS/MS) or ion-pair reverse phase HPLC (IP-RP-HPLC).
② 3' Poly(A) tail length measured by IP-RP-HPLC.
③ Product-related Impurities - dsRNA: Identified by immunoblot or ELISA.
④ Product-related Impurities - Aggregates quantified by size exclusion HPLC (SEC-HPLC).
⑤ Product-related Impurities - Fragment mRNA percentage determined by RP-HPLC.
(5) Process Rrelated Impurities
① Residual DNA template quantified by qPCR.
② Free/unbound nucleotides measured by RP–LC-MS/MS.
③ Residual T7 RNA polymerase content determined by ELISA.
(6) Potency
Target protein expression analyzed by cell-based assays.
(7) Safety
① Endotoxins complying with USP <85>.
② Bioburden adhering to USP <61>, <62>, <1115>.
(8) Other
① Appearance complying with USP <790>.
② Residual solvents complying with USP <467>.
③ pH maintained complying with USP <791>.
Note: USP<number>refers to specific chapters or guidelines in the United States Pharmacopoeia, which provide more detailed standards and testing methods.
3. Characterization and Release Testing of mRNA Drug Product
(1) Identity
① mRNA sequence identity confirmed by Sanger sequencing or Reverse transcriptase polymerase chain reaction (RT-PCR).
② Lipid identity verified by RP-HPLC with a charged aerosol detector (RP-HPLC-CAD).
(2) Content
① RNA concentration/RNA encapsulation efficiency assessed using fluorescence-based methods.
② Lipid content measured by RP-HPLC-CAD.
(3) Integrity
① LNP size and polydispersity analyzed by dynamic light scattering (DLS).
② RNA size and integrity evaluated by CGE.
(4) Purity
① Product-related Impurities - Aggregates quantified by SEC-HPLC.
② Product-related Impurities - Fragment mRNA percentage measured by IP-RP-HPLC.
(5) Potency
Expression of the target protein: Analyze using cell-based assay methods.
(6) Safety
① Endotoxin levels: Adhere to the standards outlined in USP <85>.
② Sterility testing: Conduct according to USP <71>.
(7) Other
① Physical appearance: Inspect as per USP <790>.
② Residual solvents: Assess following USP <467>.
③ Osmolarity: Measure in accordance with USP <785>.
④ Sub-visible particles: Evaluate using USP <787>.
⑤ Extractable volume: Determine according to USP <1> and USP <698>.
⑥ Integrity of container closure: Test as per USP <1207>.
⑦ pH levels: Verify following USP <791>.
Note: References such as USP <number> denote specific chapters or guidelines in the United States Pharmacopeia, detailing the standards and testing methods applicable.
Vaccine Drug Analysis Solutions
Service Advantages
1. Providing Professional and Robust Services Covering Multiple Aspects of mRNA Drug Testing
2. Offering Cost-Effective Testing with Short Experimental Periods, and Delivering Complete and Credible Reports
3. Equipping with a Variety of Large Instruments Such as High-Resolution Mass Spectrometers
Case Study
1. Stability-Indicating Ion-Pair Reverse Phase Liquid Chromatography Method for Modified mRNA
Modified mRNA is a rapidly emerging therapeutic product. Thus, developing robust stability-indicating methods to control mRNA product quality is crucial for supporting successful drug development. A paper describes an ion-pair reverse-phase liquid chromatography (IP-RPLC) method for characterizing modified mRNA exposed to various stress-inducing conditions relevant to mRNA drug development. The optimized method can be used to separate and analyze large RNAs up to 1000 nucleotides. The column temperature, flow rate of the mobile phase, and choice of ion-pair reagents were studied and optimized. Baseline separation of model RNA molecular weight standards was achieved using all the examined ion-pair reagents. The study identified that the optimized method using 100 mM triethylamine provided the highest resolution separation of the largest fragment in the ladder (750/1000 nucleotides), and also had the highest overall resolution for the selected modified mRNA compound (eGFP mRNA, 996 nucleotides). The stability-indicating efficacy of the method was demonstrated by analyzing modified eGFP mRNA upon direct exposure to heat, hydrolytic conditions and treatment with ribonucleases. The results showed that the optimized method could well monitor the degradation products formed, which appeared as shorter RNA fragments before the main peak, and assess the relative stability of mRNA under various stressed conditions.
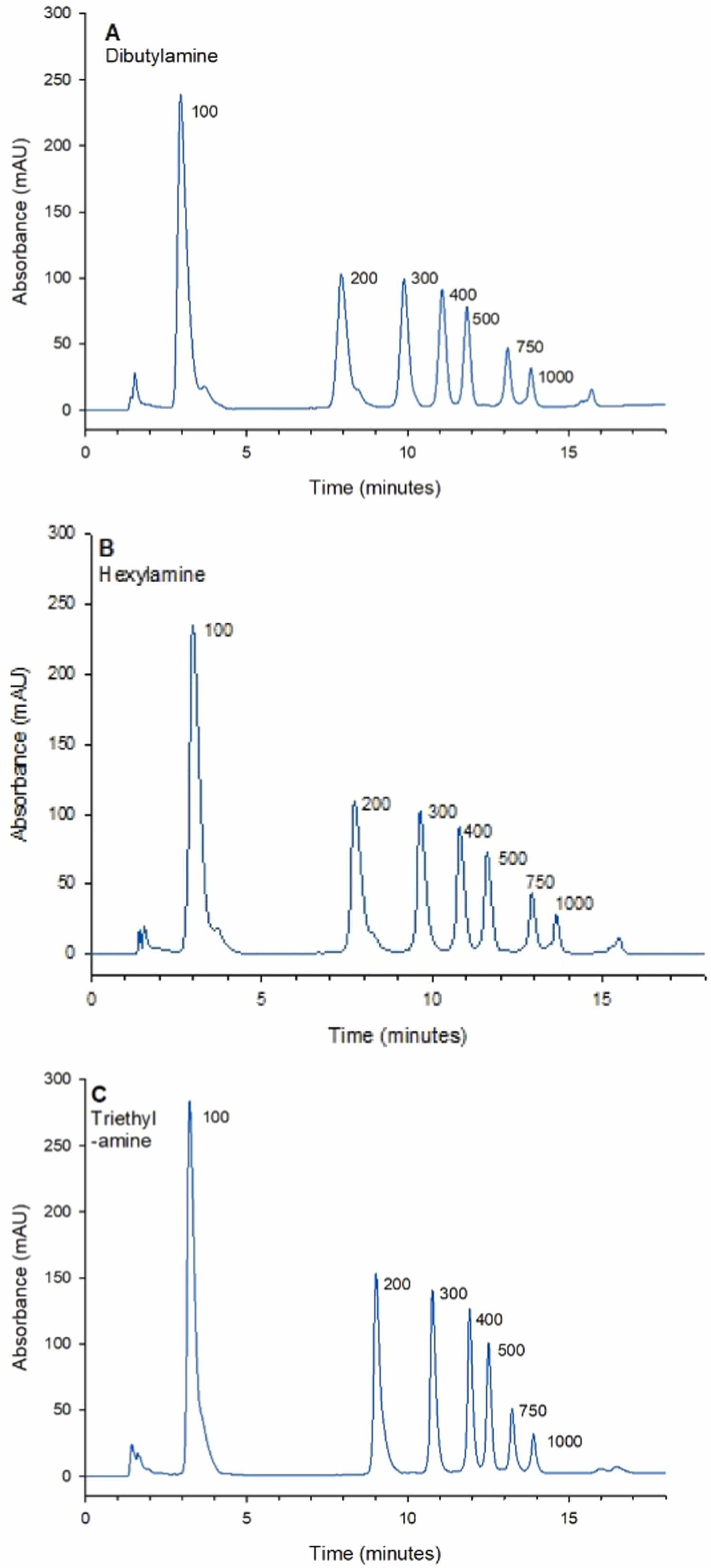
Figure 2. Comparison of Different Ion-Pair Reagents for Separating Single-Stranded RNA Molecular Weight Standards of 100-1000 Nucleotides Using the Polymer-Based DNAPac RP Column [2]
2. Anion Exchange HPLC Monitoring of mRNA In Vitro Transcription Reactions to Support mRNA Production Process Development
mRNA technology has recently proven capable of drastically reducing the timeline for developing and delivering new vaccines from years to months. The recent successful development and approval of two mRNA vaccines for COVID-19 highlighted the potential of mRNA technology for rapid vaccine development. Importantly, this RNA-based approach holds promise for therapies beyond vaccines and infectious diseases, such as cancer, metabolic disorder, cardiovascular disease, and autoimmune disease. Currently, there is a significant need to develop improved manufacturing processes to produce mRNA therapeutics with higher yield and quality. Developing suitable analytical methods for analyzing mRNA therapeutics is crucial for supporting manufacturing development and characterization of both drugs and active pharmaceutical ingredients. In this study, a high-throughput, high-efficiency HPLC workflow was developed for rapid analysis of mRNA produced using in vitro transcription (IVT). Researchers optimized anion exchange (AEX) HPLC for direct analysis of mRNA from IVT. The chromatographic analysis was completed within 6 minutes and could separate all key components in the IVT, including nucleoside triphosphates (NTPs), cap analogs, plasmid DNA, and mRNA products. Furthermore, baseline separation of NTPs was achieved, allowing accurate quantification of each NTP, which can be used to determine their consumption during the IVT reaction. Additionally, the HPLC method was used to quickly assess the purification of mRNA products, including the removal of NTPs/cap analogs and other impurities after downstream purification steps such as solid-phase extraction (SPE), oligo-dT affinity chromatography, and tangential flow filtration (TFF). Excellent accuracy was obtained using the developed method, with calibration curves of external mRNA standards and NTPs showing correlation coefficients of 0.98 and 1.0, respectively. Intraday and interday studies on the retention time stability of NTPs showed relative standard deviations of ≤0.3% and ≤1.5%, respectively. mRNA retention time variability was ≤0.13%. The method was then used to monitor the progress of the IVT reaction producing COVID spike protein (C-Spike) mRNA to measure the increase in mRNA yield and consumption of NTPs during the reaction.
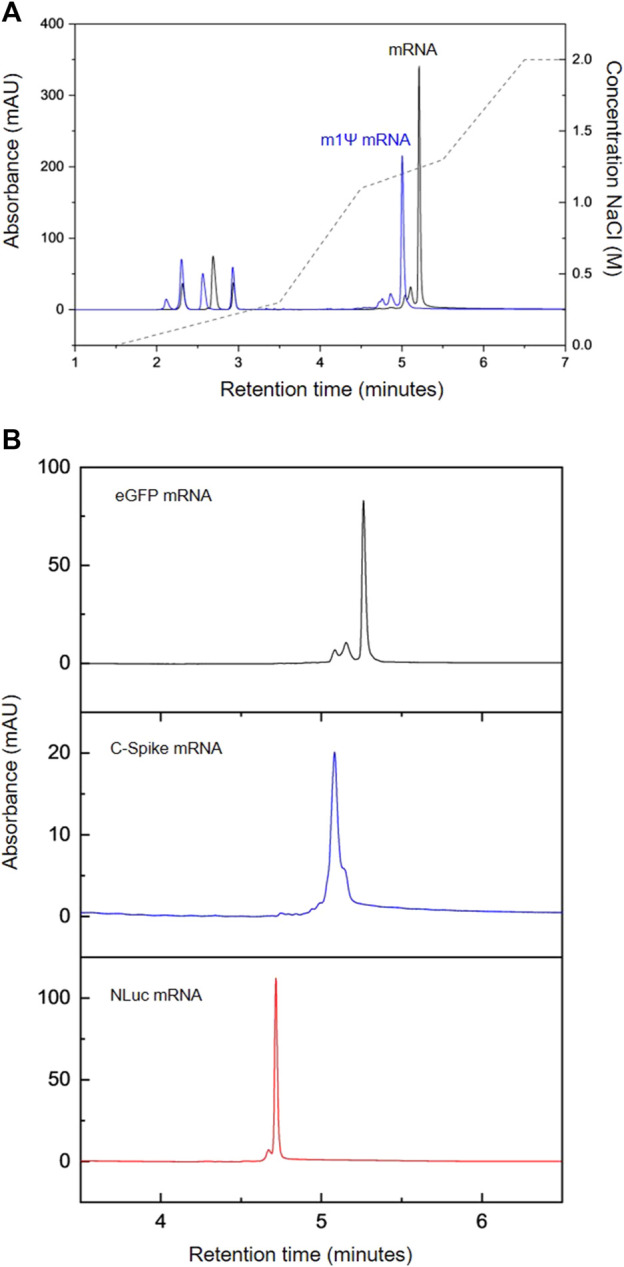
Figure 3. AEX HPLC Analysis of IVT mRNA. (A) AEX Analysis of IVT Reactions Producing Modified eGFP mRNA (m1Ψ, Blue) and Unmodified eGFP mRNA (Black). (B) AEX Analysis of a Series of Different mRNAs: eGFP (Black), C-Spike (Blue), NLuc (Red) [3]
3. Flow-NMR as a Process Monitoring Tool for mRNA IVT Reactions
Messenger RNA (mRNA)-based vaccines have played a crucial role in accelerating the end of the SARS-CoV-2 pandemic and are being actively developed as preventive measures for a range of viral diseases. The rapid adoption of mRNA-based therapeutics also leaves ample opportunities for improving mRNA-based drug development. One area of significant potential focuses on the production of mRNA drug substances, where mRNA is produced via a cell-free reaction called in vitro transcription (IVT). Process analytical technology (PAT) is an integral part of the pharmaceutical industry and is essential for facilitating agile process optimization and enhancing process quality, control, and understanding. Due to the inherent complexity and novelty of IVT reactions, effective PAT is needed to provide deep, real-time insights into the reaction process to deliver novel mRNA vaccines to patients in a faster and more cost-effective way. Here, a study showed that flow nuclear magnetic resonance (flow-NMR) could be an efficient process analysis tool for monitoring mRNA IVT reactions to support process development, optimization, and production.
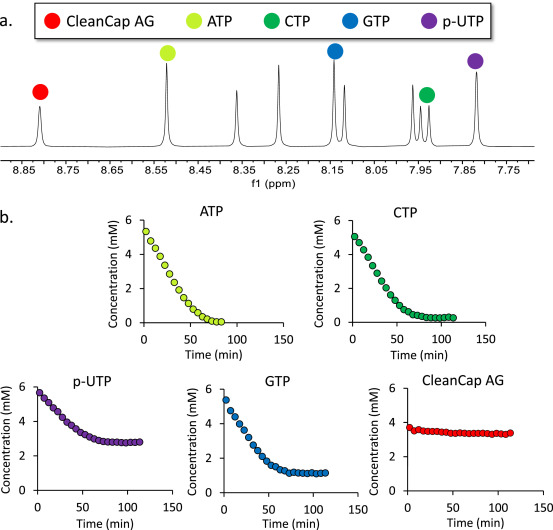
Figure 4. 1H NMR is Used to Track Changes in Four Types of Reaction NTP (ATP, GTP, CTP, and p-UTP) and Capping Reagent Cleancap AG Used in IVT Reactions [5]
4. Chip-Based Digital PCR as a Direct Quantification Method for Residual DNA in mRNA Drugs
Residual exogenous DNA, as a common contaminant in biologics, must be monitored and removed to ensure safety. dPCR technology has been widely used in DNA quantification analysis with high specificity, sensitivity, and absolute quantification. There is relatively little data support for elucidating the application of dPCR technology in residual DNA from mRNA drugs. A current study helped establish dPCR methods corresponding to two different mRNA vaccines to detect residual DNA templates. The established dPCR method had a wide linear range, good precision, accuracy, and specificity, and was not affected by encapsulation and demulsifying reagents. The method was simple, rapid, and sensitive, indicating that dPCR can also directly and accurately quantify other types of risky DNA in mRNA drugs.
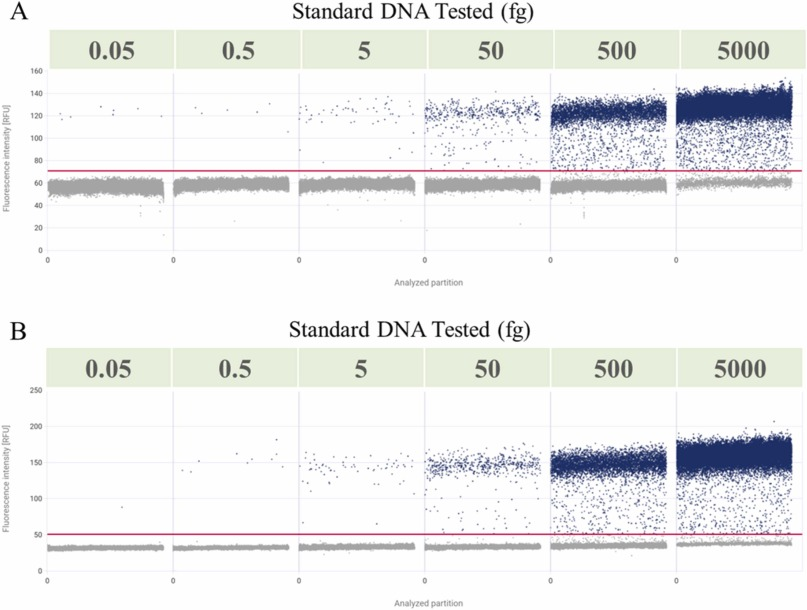
Figure 5. Linear Analysis of dPCR [5]
References
[1] Weng Y, Li C, Yang T, Hu B, Zhang M, Guo S, Xiao H, Liang XJ, Huang Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol Adv. 2020 May-Jun;40:107534. doi: 10.1016/j.biotechadv.2020.107534. Epub 2020 Feb 21. PMID: 32088327.
[2] Currie J, Dahlberg JR, Lundberg E, Thunberg L, Eriksson J, Schweikart F, Nilsson GA, Örnskov E. Stability indicating ion-pair reversed-phase liquid chromatography method for modified mRNA. J Pharm Biomed Anal. 2024 Apr 10;245:116144. doi: 10.1016/j.jpba.2024.116144. Epub ahead of print. PMID: 38636193.
[3] Welbourne EN, Loveday KA, Nair A, Nourafkan E, Qu J, Cook K, Kis Z, Dickman MJ. Anion exchange HPLC monitoring of mRNA in vitro transcription reactions to support mRNA manufacturing process development. Front Mol Biosci. 2024 Mar 7;11:1250833. doi: 10.3389/fmolb.2024.1250833. PMID: 38516194; PMCID: PMC10955092.
[4] Sarkar A, Dong G, Quaglia-Motta J, Sackett K. Flow-NMR as a Process-Monitoring Tool for mRNA IVT Reaction. J Pharm Sci. 2024 Apr;113(4):900-905. doi: 10.1016/j.xphs.2023.11.021. Epub 2023 Nov 24. PMID: 38008177.
[5] Fan W, Zhao L, Yu L, Zhou Y. Chip-based digital PCR as a direct quantification method for residual DNA in mRNA drugs. J Pharm Biomed Anal. 2024 Jan 20;238:115837. doi: 10.1016/j.jpba.2023.115837. Epub 2023 Oct 31. PMID: 37952451.
How to order?







