Small Molecule Drug Target Identification and Validation Service
In modern drug discovery and development processes, drug targets and mechanisms of action remain the two biggest challenges. The correct drug target not only relates to the drug's efficacy and specificity but also involves drug safety and potential side effects. Drug targets refer to the direct binding sites of drugs with endogenous biomacromolecules in the body, mainly including receptors, enzymes, ion channels, transporters, nucleic acids, and other biomacromolecules. Identifying drug action targets is crucial for the design of new drugs. It helps us understand the mechanism of drug action, establish the connection between drug activity and biological functions, predict possible side effects and resistance mechanisms, and discover new therapeutic targets. Through target identification, we can ensure the efficiency of the drug development process and the safety of the drugs after they are marketed.
New drug action targets are breakthroughs in a series of new drug discoveries. Searching for new drug action targets has become a focal point in the fierce competition of innovative drug research and development. From the perspective of drug development, the effect of a drug largely depends on its target. Currently, two main drug discovery strategies are used: phenotype-based drug discovery and target-based drug discovery. Phenotype-based drug discovery involves screening small molecules or peptides in cells, tissues, or organs based on existing pharmacology. Target-based drug discovery initially involves identifying targets and then recognizing active molecules. With the rapid development of molecular biology, the paradigm of target-based drug discovery has replaced the traditional phenotype-based methods because it can enhance screening capabilities and define reasonable drug discovery schemes.
MtoZ Biolabs relies on a high-resolution mass spectrometry platform, combined with affinity chromatography and active site-directed probe technologies, to develop and validate a variety of target-based small molecule drug target identification platforms, providing you with a full process of molecular interaction identification research services, including protein-protein interactions (PPI), protein-small molecule drug interactions, which can accurately identify and validate small molecule drug targets, assisting in the discovery and development of innovative drugs. MtoZ Biolab's target-based small molecule drug target identification scheme include:
Based on ABPP Strategy Small Molecule Drug Target Identification and Validation
Activity-based protein profiling (ABPP) combines activity-based probes with proteomics techniques to identify small molecule protein targets, even the active sites of target proteins. ABPP utilizes activity site-directed chemical probes composed of two elements: (1) an active site-directed reactive group used to bind and covalently label a specific subset (or family) of catalytically related enzymes, (2) a reporting label (e.g., fluorescent group, biotin, acetylene or azide) used for detection/quantification and/or enrichment/identification of labeled enzymes. In principle, the active groups of small molecules interact directly with the target protein and reporting groups to facilitate target capture. Depending on the selected reporting group, different follow-up experiments can be conducted. For example, fluorescent groups can be used for rapid gel screening and identification of small molecules in cells or animals, and biotin can be used for protein enrichment, followed by the identification of target protein by MS.
Wang, S. et al. Front Pharmacol. 2018.
Figure 1. Workflow of Activity-Based Protein Analysis
1. Services at MtoZ Biolabs
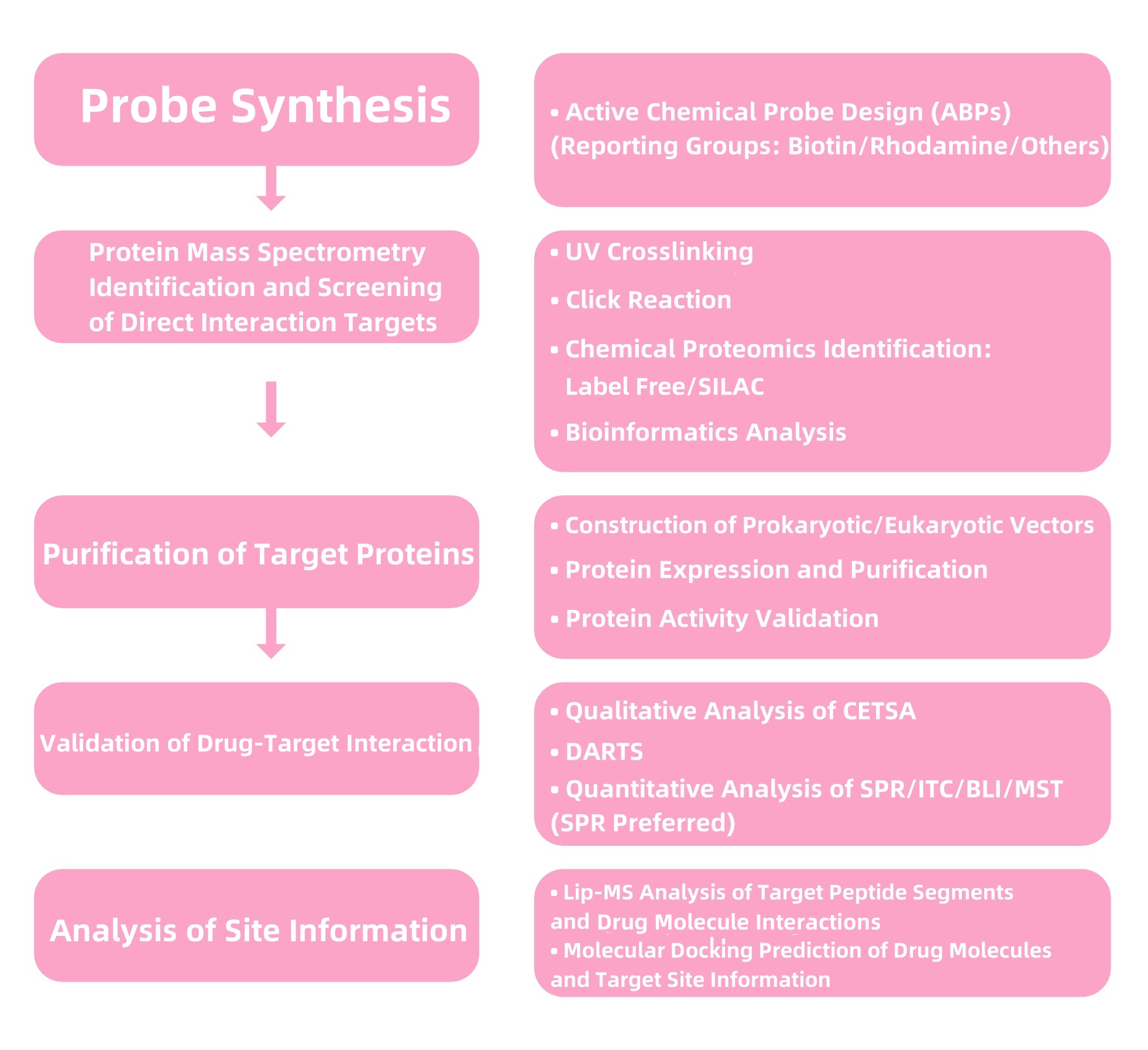
Figure 2. Service Content
2. Service Advantages
(1) Specificity
Highly specific recognition of active proteins by specifically labeling active enzymes with specific activity probes.
(2) Functional analysis
Direct assessment of the enzyme activity state of proteins, not just their expression level or structural changes.
(3) Real-time monitoring
Monitor the activity of enzymes under physiological conditions within cells, capturing the immediate impact of drugs.
(4) Non-invasive marking
The covalent binding of the probe to the protein is non-invasive. It does not interfere with normal function of cells, and maintains the physiological activity of the biological samples.
(5) Wide applicability
ABPP technology can be used for cell lysates, live cells, animal lysates, and even living animals.
Based on Proteome-Wide Thermal Stability Profiling Strategy Small Molecule Drug Target Identification
Chang, J. et al. Nat Prod Rep. 2016.
Figure 3. Workflow of Small Molecule Drug Target Identification Based on Proteome-Wide Thermal Stability Profiling Strategy
1. Services at MtoZ Biolabs
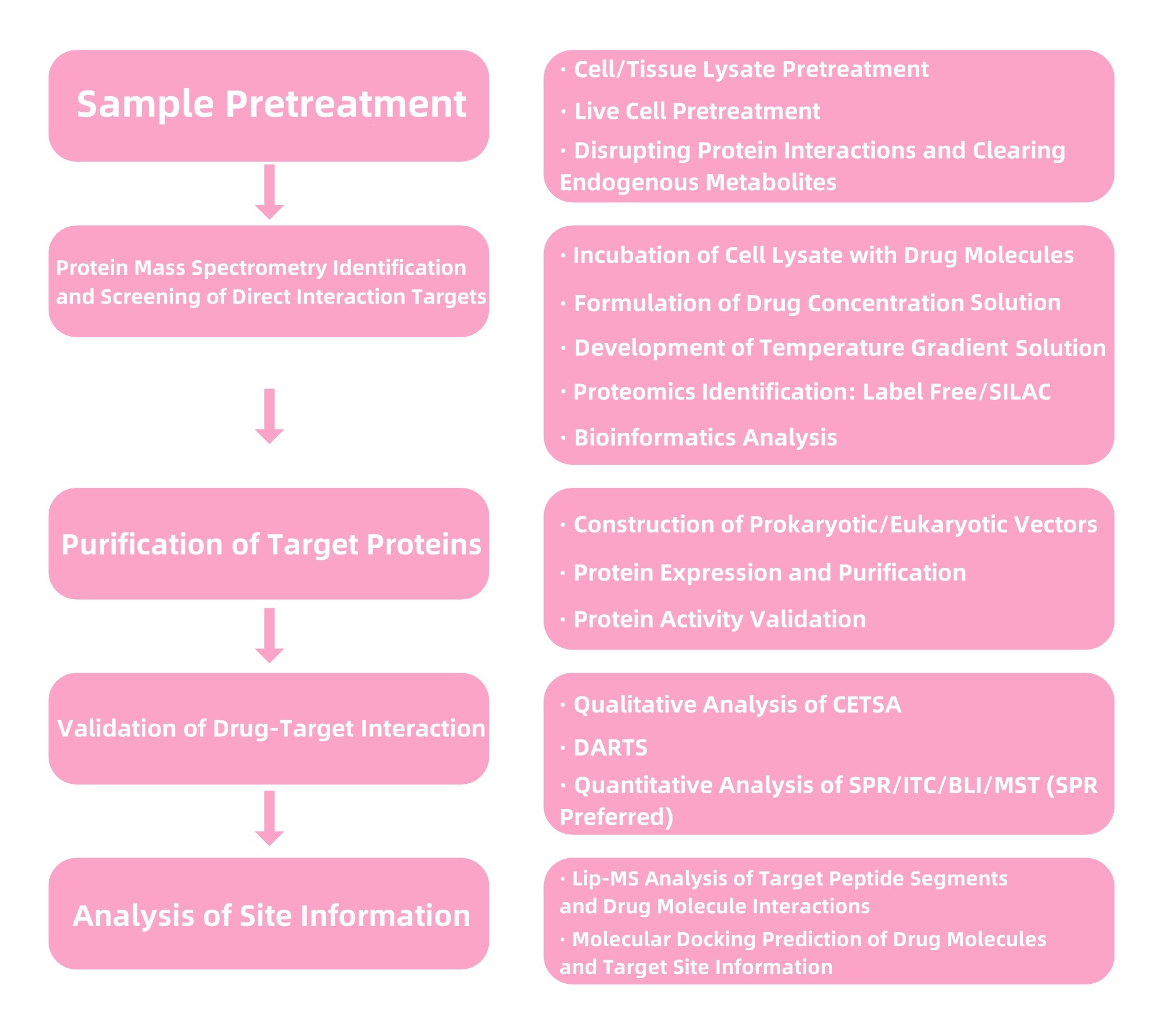
Figure 4. Service Content
2. Service Advantages
(1) Global and unbiased
Explore the interactions between small molecules and proteins at the proteome level without bias.
(2) In-cell action analysis
Capture real drug-protein interactions in the physiological environment of cells, reflecting the actual action of drugs.
(3) Quantitation
Provide quantitative information about the strength and stability of drug-target interactions.
(4) Reliability
Based on LiP-MS Strategy Small Molecule Drug Target Identification
Limited proteolysis-mass spectrometry (LiP-MS) combines protein limited proteolysis and MS techniques to identify protein fragments generated by limited cleavage of proteins by proteases under specific conditions are further identified by MS analysis. In the process of small molecule drug target identification, small molecule drugs are incubated with their potential protein targets. The drug molecules bind to the proteins and cause a change in the protein conformation, which affects the proteolytic pattern of the proteins. By comparing the proteolytic fragments of proteins under drug-treated and untreated conditions, the protein regions interacting with the drug can be identified, thereby inferring the drug's action target. This is extremely significant for analyzing the mechanism of drugs and discovering new drugs, especially in targeted drug molecule design and optimizing drug efficacy.
Piazza. et al. Cell. 2018.
Figure 5. Workflow of Small Molecule Drug Target Identification Based on LiP-MS Strategy
1. Services at MtoZ Biolabs
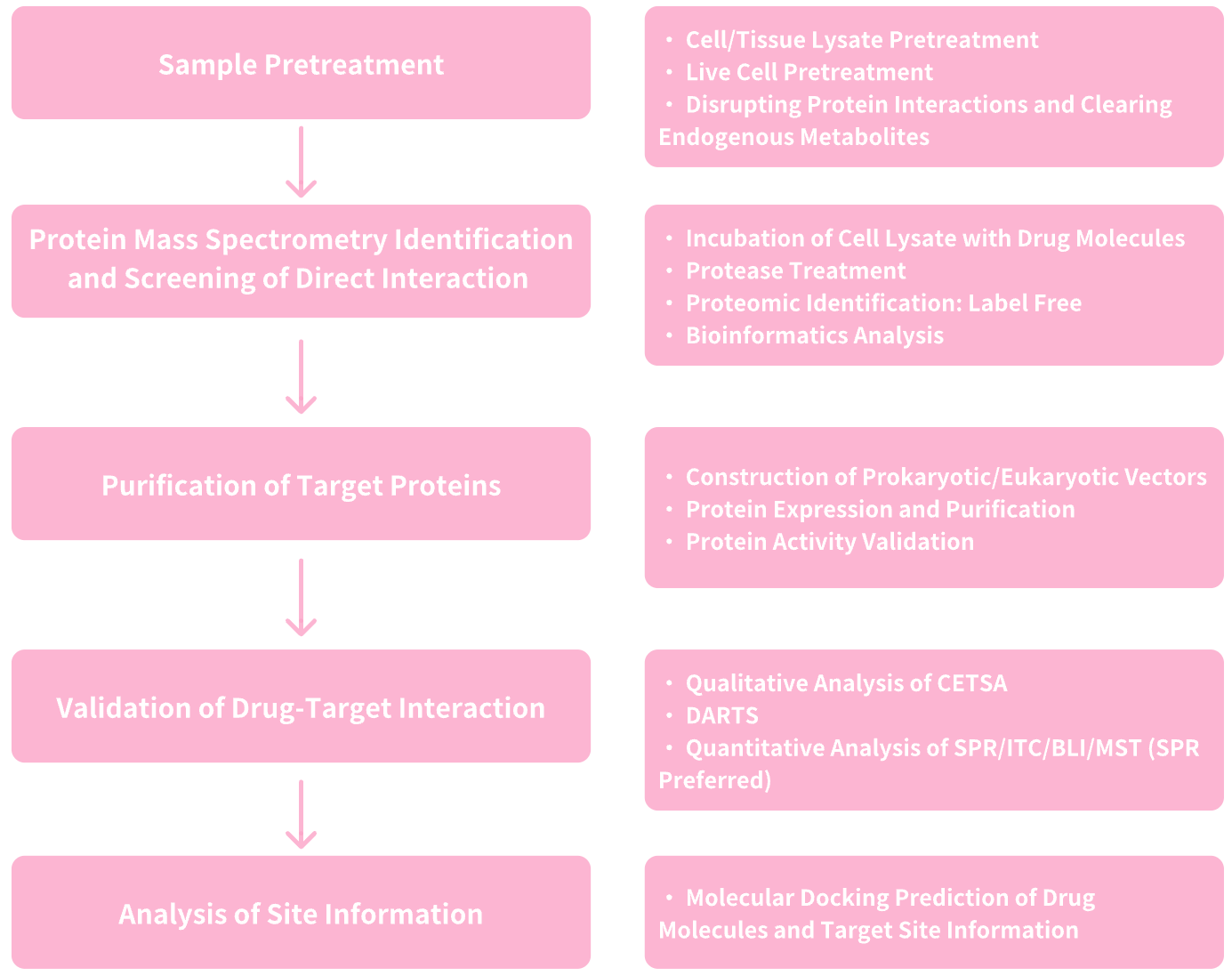
Figure 6. Service Content
2. Service Advantages
(1) High throughput and sensitivity
LiP-MS can analyze thousands of protein samples, even those low-abundance protein targets can be identified.
(2) No prior knowledge required
This method does not require prior information about potential targets and can discover unknown drug action sites in a wide range of proteins.
(3) Applicability in vitro and in vivo
LiP-MS can be conducted both in vitro and in vivo, providing more information about how drugs interact with targets.
(4) Structural dynamics analysis
LiP-MS can provide a dynamic process of protein structural changes after binding with small molecules.
Based on DARTS Strategy Small Molecule Drug Target Identification
Drug affinity responsive target stability (DARTS) is a relatively fast and direct method to identify potential protein targets of small molecules, relying on the protection from proteolysis provided by the interaction with small molecules. DARTS simply treats equal aliquots of cell lysates with the compound of interest and a control or inactive analog, followed by limited digestion with a protease. The samples are then separated by SDS-PAGE and stained to identify protein bands protected from small molecule proteolysis. Finally, MS is used to identify the proteins in each band. This unbiased method has been successfully used to identify new protein targets of natural products and other bioactive small molecules.
Chang, J. et al. Nat Prod Rep. 2016.
Figure 7. Workflow of Small Molecule Drug Target Identification Based on DARTS Strategy
1.Services at MtoZ Biolabs
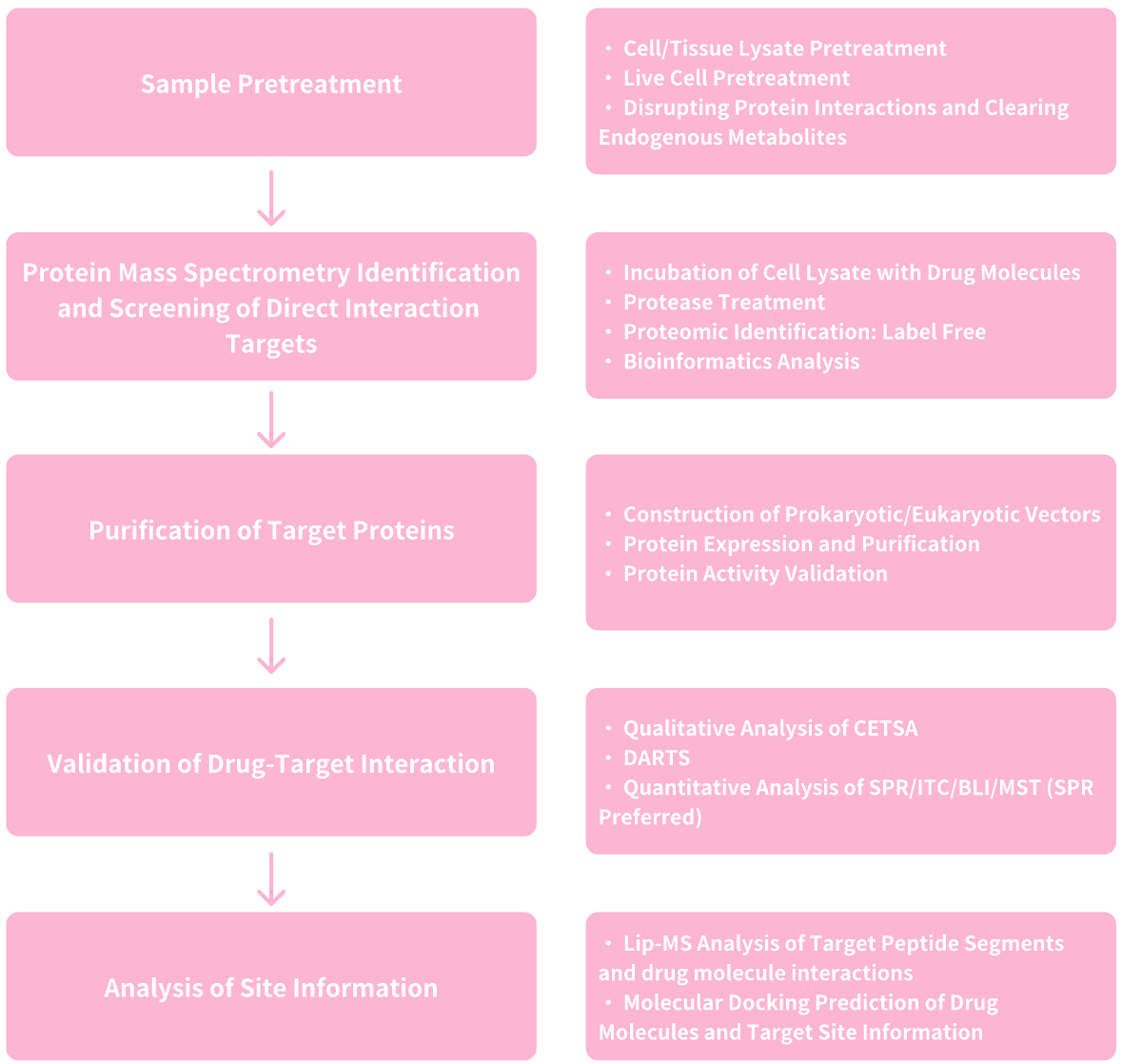
Figure 8. Service Content
2. Service Advantages
(1) Non-labeling
The biggest advantage of DARTS is that it can use natural small molecules without fixing or modifying drugs or proteins (e.g., incorporating biotin, fluorescence, radioactive isotopes), thereby avoiding potential effects on drug activity or protein structure.
(2) Directness
The experiment can be directly conducted in complex biological samples, does not involve protein purification, making the data closer to physiological state.
(3) Fidelity
The experiment can preserve the physiological interaction between drugs and their potential targets, providing more realistic drug-target interaction information.
(4) Applicability
The experiment can be used for any type of small molecule drug, regardless of its chemical properties, and has been widely used to trace proteins from animals, fungi, bacteria, and plants.
(5) Timeliness and cost-effectiveness
The experimental steps are relatively simple and quick, and the results can be obtained in a short time. The experiment is more economical and does not require expensive equipment and reagents.
Case Study
1. Identification of the Direct Action Target of the Anti-diabetic Drug Metformin Using ABPP Technology
Ma, T. et al. Nature. 2022.
Figure 9. Identification of Metformin Interaction Proteins Through a Metformin Probe
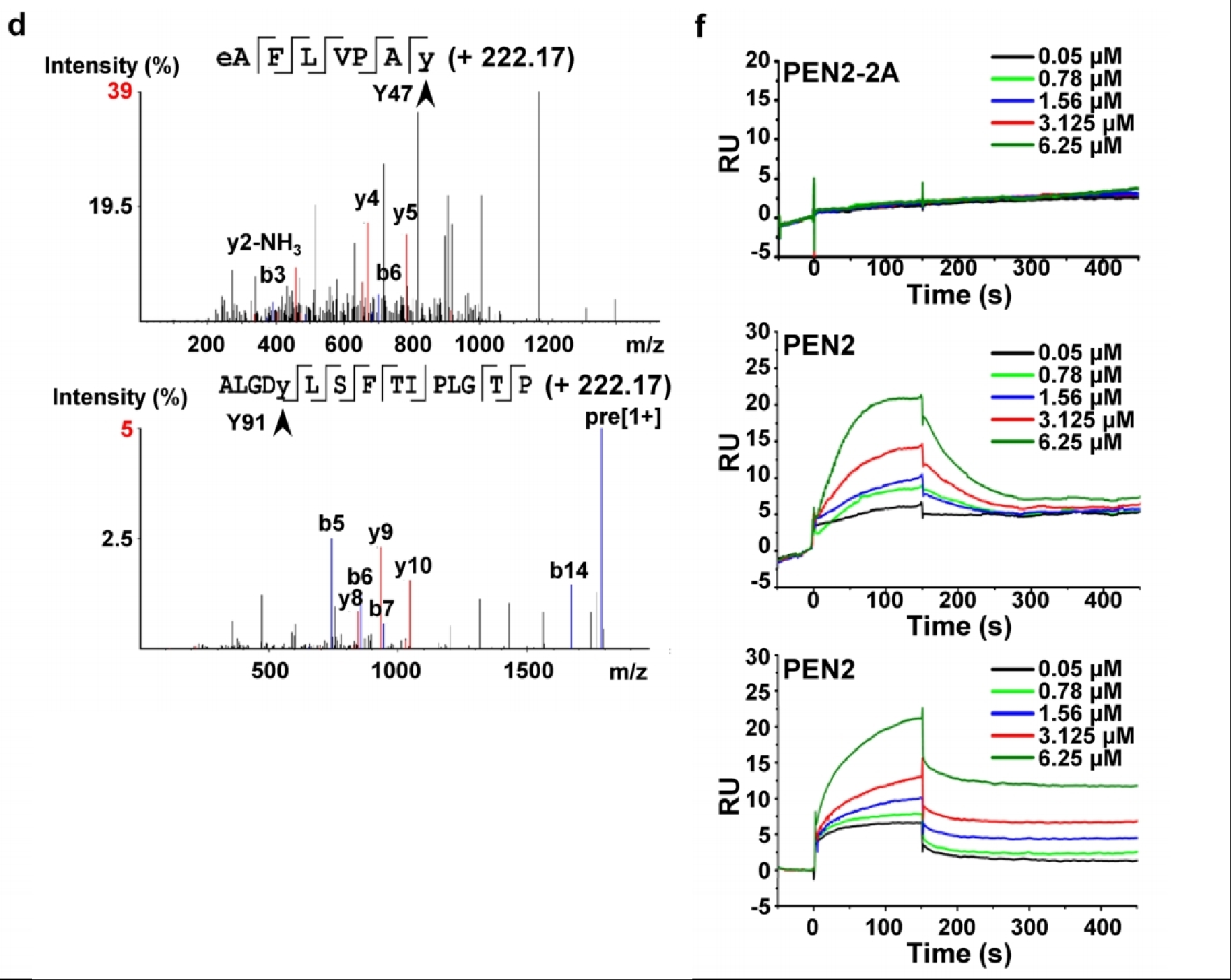
Ma, T. et al. Nature. 2022.
Figure 10. Using Mass Spectrometry to Determine the Binding Site of PEN2 with Metformin, and Then Analyzing Its Interaction with Metformin Through SPR Analysis
2. Comprehensive Characterization of Ligand-induced Changes in the Abundance and Thermal Stability of Plasma Membrane Proteins Using Cell Surface Proteome-wide Thermal Stability Profiling to Study Drug Binding with Extracellular Receptors and Transport Proteins
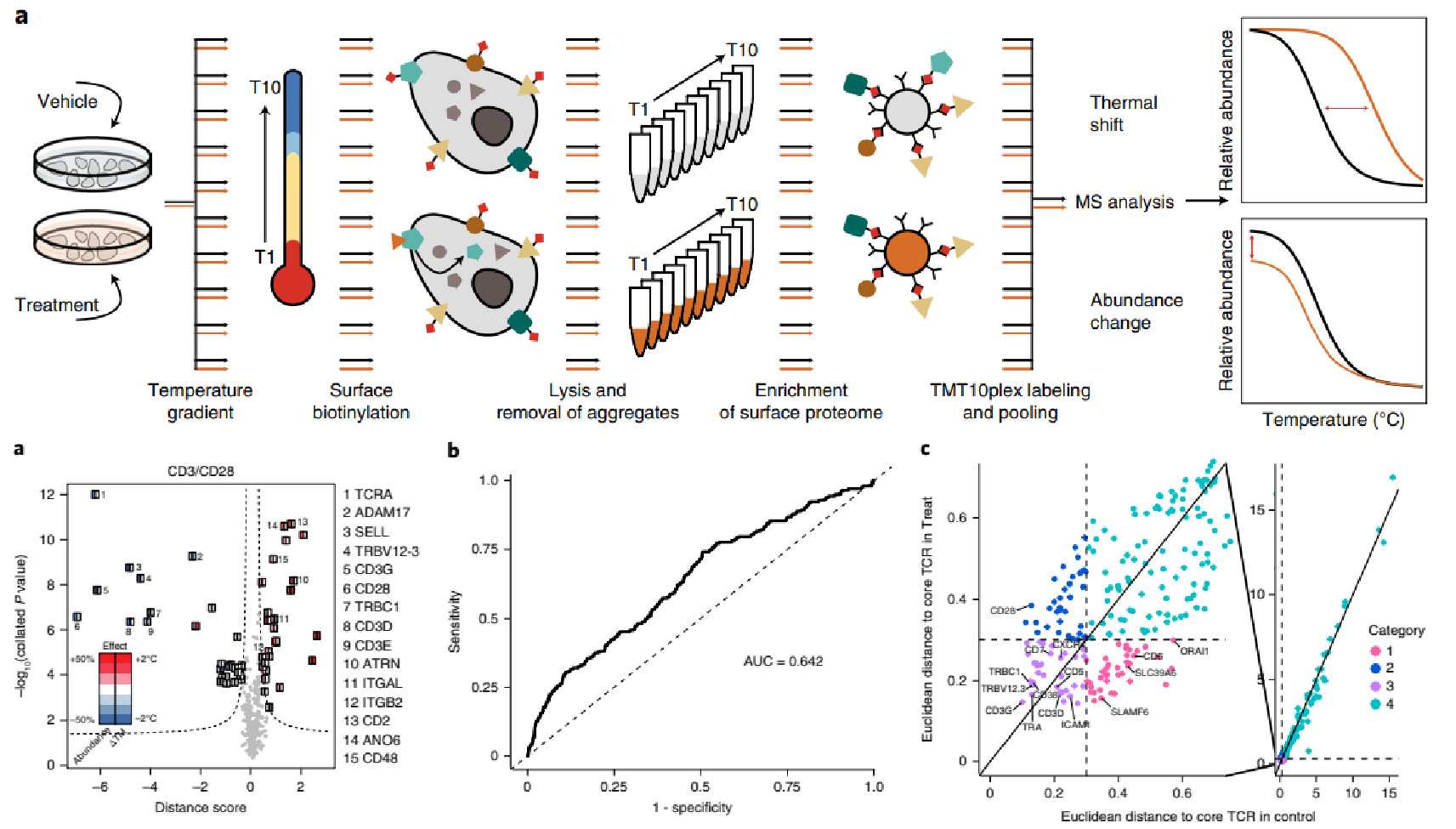
Kalxdorf, M. et al. Nat Methods. 2021.
Figure 11
3. Screening Cerebrospinal Fluid for Aging-related New Proteins and Complexes Using LiP-MS
Shuken, S. R. et al. Nat Aging. 2022.
Figure 12
4. Revealing Actin as the Biological Target of Anti-cancer Flavonoid Drug Artemisinin Using DARTS Technology
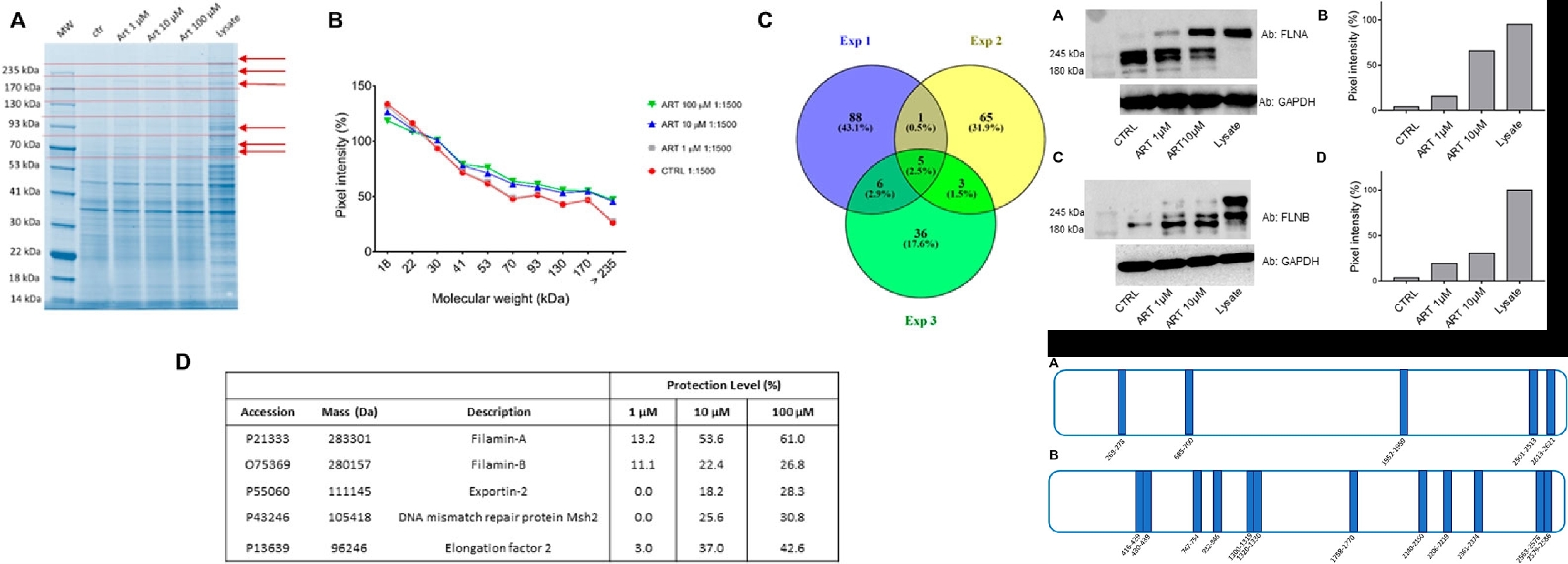
Ferraro, G. et al. Front Mol Biosci. 2022.
Figure 13
How to order?







