Peptidomics-Based PROTAC Research Service
PROTAC (Proteolysis Targeting Chimera) is a class of small molecules that induce selective degradation of target proteins via the ubiquitin-proteasome system (UPS). By linking target protein ligands with E3 ubiquitin ligase ligands, PROTACs promote the degradation of specific proteins, enabling "targeted removal" rather than "functional inhibition."
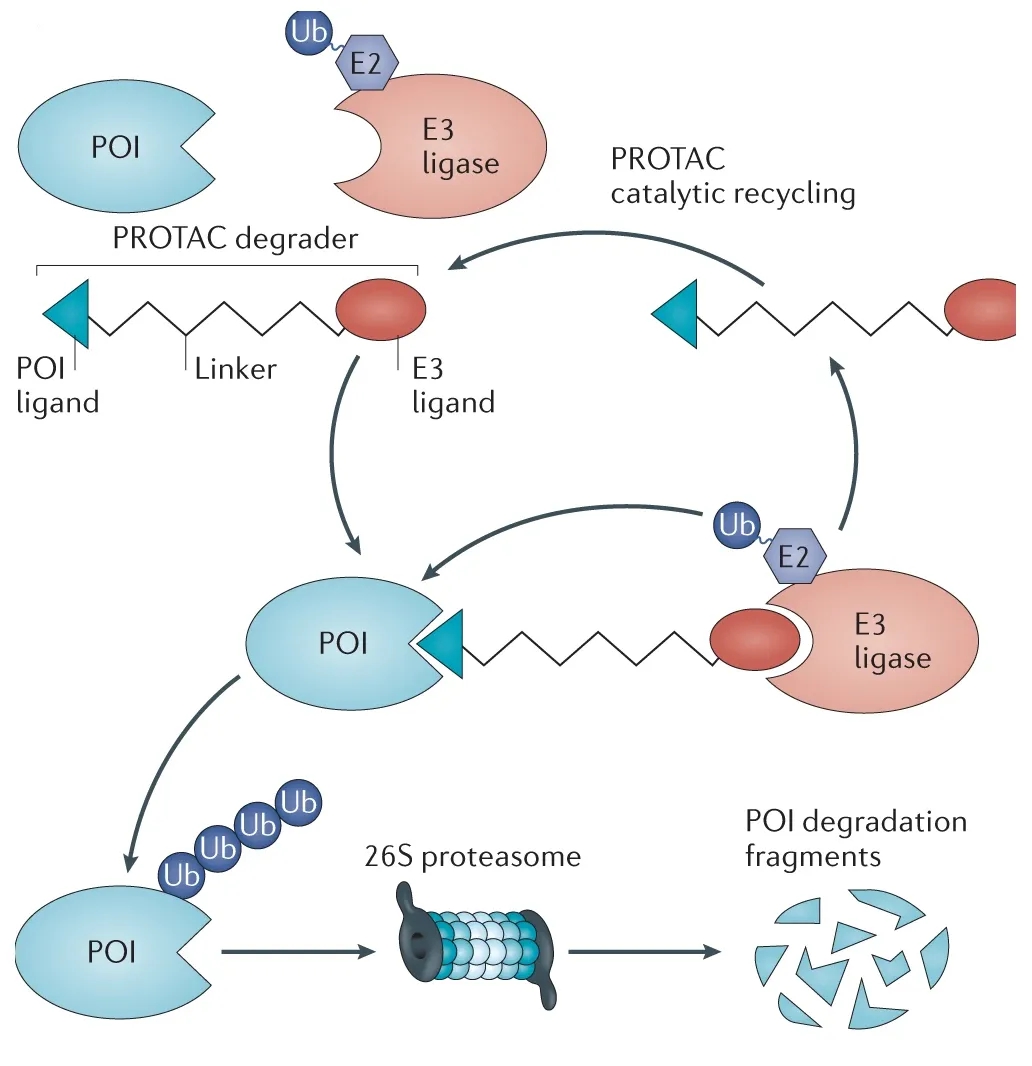
Dale, B. et al. Nature Reviews Cancer, 2021.
Figure 1. Mechanism of PROTAC-Mediated Target Protein Degradation
In understanding PROTAC mechanisms and validating targets, peptidomics offers a critical complement. This high-throughput approach identifies protein degradation products, immune-related peptides, and small peptides in signaling pathways, providing insights into degradation events, immune responses, and off-target effects. MtoZ Biolabs offers Peptidomics-Based PROTAC Research Service that combines advanced mass spectrometry platforms with extensive peptidomics expertise to provide comprehensive experimental support and data interpretation for protein degradation studies.
Technical Principles
Peptidomics-Based PROTAC research relies on high-resolution LC-MS/MS technology to systematically analyze endogenous low-molecular-weight peptides from cells, tissues, or body fluids. This approach identifies protein degradation products, immunogenic peptides, and small signaling peptides associated with PROTAC treatment. By comparing peptide profiles across different treatment groups and combining functional annotation with structural prediction, we can indirectly confirm degradation targets, identify potential immunogenic fragments, and support PROTAC mechanism optimization and target validation.
Services at MtoZ Biolabs
Our Peptidomics-Based PROTAC Research Service covers several key modules that can be customized based on your research objectives:
· Endogenous Degradation Peptide Analysis
Identify and trace protein cleavage peptides resulting from PROTAC treatment, verifying target protein degradation and elucidating cleavage patterns.
· Immune Peptide Presentation Profiling
Track changes in peptides presented via the MHC-I pathway, assessing whether PROTAC-induced degradation triggers immune responses, supporting immunotherapy research.
· Peptides Involved in Signal Regulation
Uncover small peptides related to cellular stress and transcriptional regulation, revealing downstream effects of PROTAC.
· Peptide Biomarker Discovery
Discover biomarkers linked to PROTAC efficacy or off-target effects, facilitating further investigation of therapeutic potential.
Analysis Workflow
MtoZ Biolabs’ Peptidomics-Based PROTAC Research Service includes the following key steps to ensure the most valuable research data for our clients:
1. Sample Processing and Peptide Extraction
Extract endogenous low molecular weight peptides from cells, tissues, or biofluids, removing larger protein background to ensure purity.
2. LC-MS/MS Analysis
Use high-efficiency Nano-LC combined with Orbitrap mass spectrometry to achieve precise peptide detection.
3. Quantitative Analysis and Comparison
Utilize labeled or label-free quantification to compare peptide abundance changes between PROTAC-treated and control groups.
4. Immune Peptide and Degradation Product Identification
Identify MHC-presented peptides, target-derived peptides, or functionally relevant peptides to validate degradation efficiency and pathway mechanisms.
5. Bioinformatics Analysis and Structural Prediction
Integrate database searches, GO/KEGG functional annotations, and structural domain recognition for biological interpretation.
6. Results Reporting
Provide a comprehensive report with quantitative tables, differential analysis, functional annotations, and PTM details, ensuring clarity and relevance to your research.
Why Choose MtoZ Biolabs?
✔ Cutting-Edge Instrumentation: Our lab is equipped with the latest Orbitrap Fusion Lumos and Nano-LC systems for precise peptide identification and quantification.
✔ Tailored Research Plans: We design customized experiments and analyses based on your unique research needs, offering flexible options for each project.
✔ Comprehensive Data and Reports: Along with raw data, we provide thorough analysis reports with scientific interpretations to ensure seamless progress in your research.
✔ Fast and Efficient Service: We ensure timely updates and quick turnaround times to keep your project on track and meet deadlines.
✔ One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
FAQ
Q1: How do PTMs of peptides affect PROTAC research?
PTMs can alter peptide stability, immune-presentation, or interaction with degradation pathways. MtoZ Biolabs detects common PTMs such as phosphorylation and acetylation to assess their impact on PROTAC-induced degradation and off-target risks.
Q2: What are the sample submission requirements?
We accept samples including cells (≥10⁷ cells), tissue (≥50 mg), and protein extracts (≥1 μg/μL). Detailed submission guidelines are available upon request to ensure your samples meet the required standards.
Q3: What is the difference between Peptidomics and Proteomics in PROTAC research?
Peptidomics focuses on naturally occurring short peptides (<10 kDa), directly reflecting protein degradation products. Proteomics, however, analyzes full-length protein abundance changes. Peptidomics offers a complementary method in PROTAC research for quickly confirming degradation events and monitoring immune peptides.
MtoZ Biolabs is committed to providing specialized, systematic support for protein degradation research. Our Peptidomics-Based PROTAC Research Service complements proteomics by helping researchers investigate degradation mechanisms, validate target engagement, and evaluate off-target effects and immune responses. If you are developing or validating PROTAC candidates, feel free to contact us for customized technical support and analysis solutions.
Related Services
Custom PROTAC Synthesis Service
PROTAC Drug Off-Target Protein Assessment Service
Peptidomics-Based Research Service
Peptidomics-Based Proteolytic Pathways Research Service
How to order?







