Protein Lipidation Analysis Service
Protein lipidation is a vital post-translational modification (PTM) involving the covalent attachment of lipid moieties to protein substrates. This modification enhances membrane anchoring, regulates subcellular localization, and modulates protein activity. It plays essential roles in signal transduction, vesicle trafficking, immune regulation, and disease progression. Common lipidation types include N-acylation (e.g., myristoylation), S-acylation (e.g., palmitoylation), prenylation, GPI anchoring, and cholesterol modification, each with distinct substrate selectivity and functional impact. Due to the low abundance, high heterogeneity, and dynamic nature of lipidation, advanced analytical technologies are required for precise identification and functional interpretation.
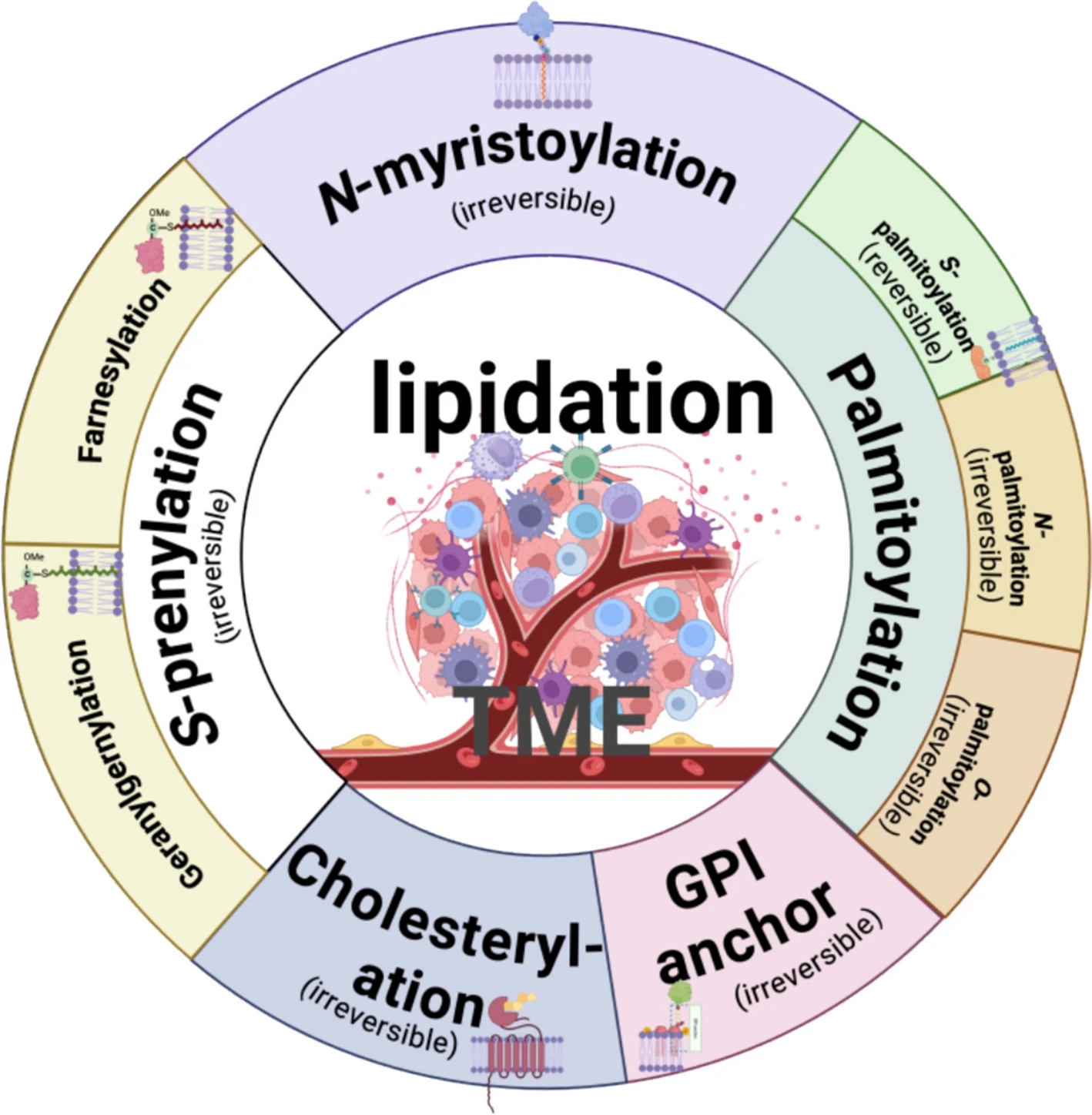
Xu, M. et al. Mol Cancer. 2025.
Figure 1. Major Types of Protein Lipidation
MtoZ Biolabs provides a comprehensive Protein Lipidation Analysis Service powered by high-resolution Orbitrap mass spectrometry, targeted enrichment strategies, and robust bioinformatics pipelines. Our platform supports the discovery of lipid-modified proteins, accurate site localization, condition-specific quantification, and functional insights into lipidation-driven regulation.
Services at MtoZ Biolabs
1. Targeted Protein Lipidation Analysis
MtoZ Biolabs offers targeted analysis of lipidation on client-specified proteins. Clients may submit one or multiple target proteins for precise identification and quantification of lipidation sites. Following proteolytic digestion and peptide-level enrichment, high-resolution LC-MS/MS is used to deliver accurate site-level characterization of lipid modifications.
2. Lipidation Proteomics
For comprehensive studies, MtoZ Biolabs provides lipidation proteomics to profile lipid-modified proteins across complex biological samples. Using large-scale LC-MS/MS with optimized peptide enrichment workflows, this service enables global mapping and quantitative comparison of lipidation patterns across different biological states or treatment conditions.
Analytical Approach
To meet diverse research goals, our Protein Lipidation Analysis Service employs three complementary and powerful technologies:
· ABE-MS (Acyl-Biotin Exchange Mass Spectrometry)
Ideal for detecting S-palmitoylation, this approach uses thiol blocking, hydroxylamine cleavage, biotin tagging, and affinity enrichment to enable confident site-specific identification via mass spectrometry.
· CR-MS (Click Chemistry-based Mass Spectrometry)
Combines metabolic labeling with click chemistry to identify S-prenylation and related modifications. This method supports dynamic lipidation profiling and fluorescent imaging.
· iFAT-MS (Isotope-coded Fatty Acid Translocase Mass Spectrometry)
Enables structural and quantitative analysis of fatty acid modifications through methyl esterification and stable isotope labeling, revealing lipidation stoichiometry and structural features.
These techniques can be used individually or in combination to accommodate both targeted and proteome-wide lipidation analyses.
Analysis Workflow
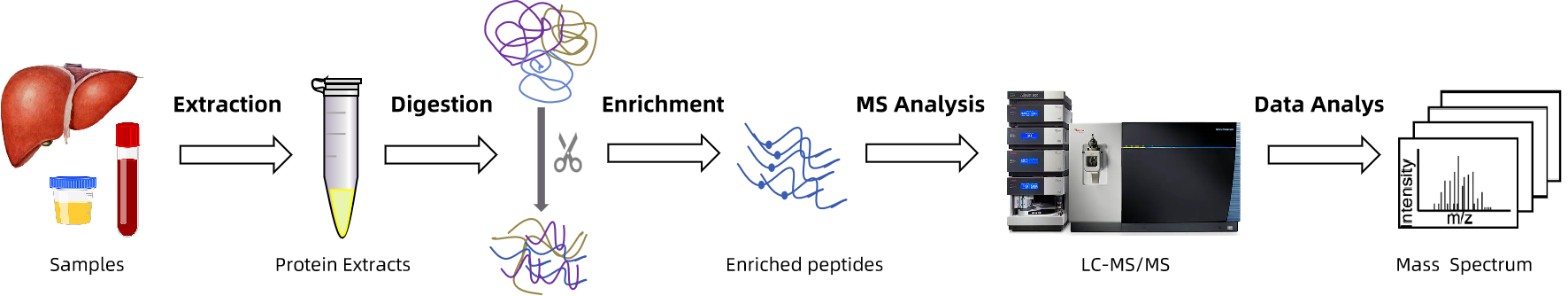
MtoZ Biolabs implements a standardized protein lipidation analysis pipeline to ensure data accuracy, reproducibility, and interpretability:
1. Sample Preparation and Protein Extraction
Tailored lysis protocols are applied to preserve lipidation integrity across various sample types.
2. Lipidated Peptide Enrichment
Enrichment is performed using ABE-MS, CR-MS, or iFAT-MS depending on the modification type and study objective.
3. Nano-LC-MS/MS Analysis
High-resolution mass spectrometry is conducted using Orbitrap Fusion Lumos or Q Exactive HF systems.
4. Bioinformatics Analysis
Includes lipidation site identification, differential quantification, GO/KEGG pathway analysis, motif discovery, subcellular localization, domain prediction, and interaction network analysis.
Why Choose MtoZ Biolabs?
✅ High-Resolution MS Platforms
Utilization of Orbitrap Fusion Lumos and other high-end instruments ensures precise detection and site-level localization.
✅ Extensive Lipidation Type Coverage
Supports profiling of N-acylation, S-palmitoylation, prenylation, GPI anchoring, and cholesterol-related modifications.
✅ Flexible Enrichment Approaches
Leverages ABE-MS, CR-MS, and iFAT-MS to suit diverse lipidation forms and sample contexts.
✅ Versatile Quantitative Strategies
Compatible with both TMT/iTRAQ and label-free quantification for comparative and time-resolved studies.
✅ Transparent Pricing and Customization
Clear fee structure with no hidden costs. Offers both standardized and fully tailored project solutions.
Sample Submission Suggestions
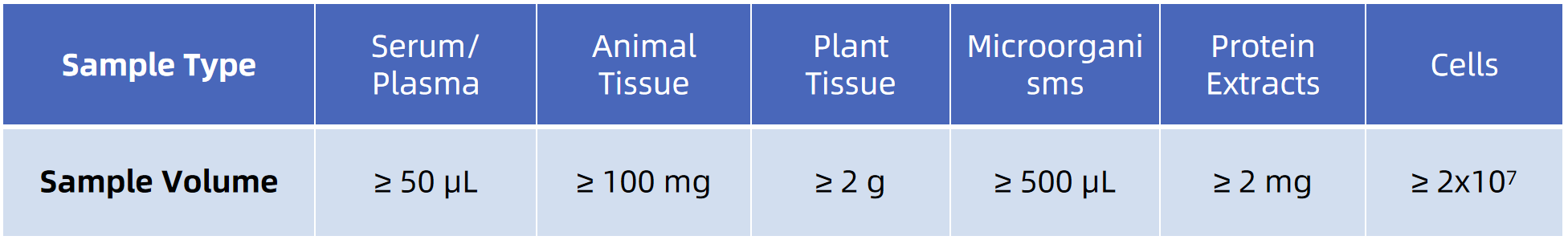
· Store samples at –80°C; avoid repeated freeze-thaw cycles.
· For samples with special treatment or buffer systems, please contact us in advance.
· A detailed sample preparation guide is available through MtoZ Biolabs technical support.
Applications
· Membrane Protein Characterization: Identify and characterize lipidation of membrane-associated proteins and elucidate anchoring mechanisms.
· Signal Transduction and Protein Localization: Investigate the role of lipidation in modulating signal protein activity and spatial distribution.
· Disease Mechanism Studies: Reveal lipidation changes in cancer, neurodegeneration, and other disease states.
· Drug Target Discovery: Discover and validate lipidated proteins or modifying enzymes as therapeutic targets.
· Protein Engineering: Guide rational design strategies for engineering lipidation in synthetic proteins.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
Whether your goal is to decode lipidation dynamics in cellular regulation or build comprehensive modification maps for translational research, MtoZ Biolabs is your trusted partner. Our Protein Lipidation Analysis Service combines cutting-edge instrumentation, expert scientific interpretation, and personalized support to accelerate your research and enable impactful discoveries.
Related Services
N-myristoylation Proteomics Service
How to order?







