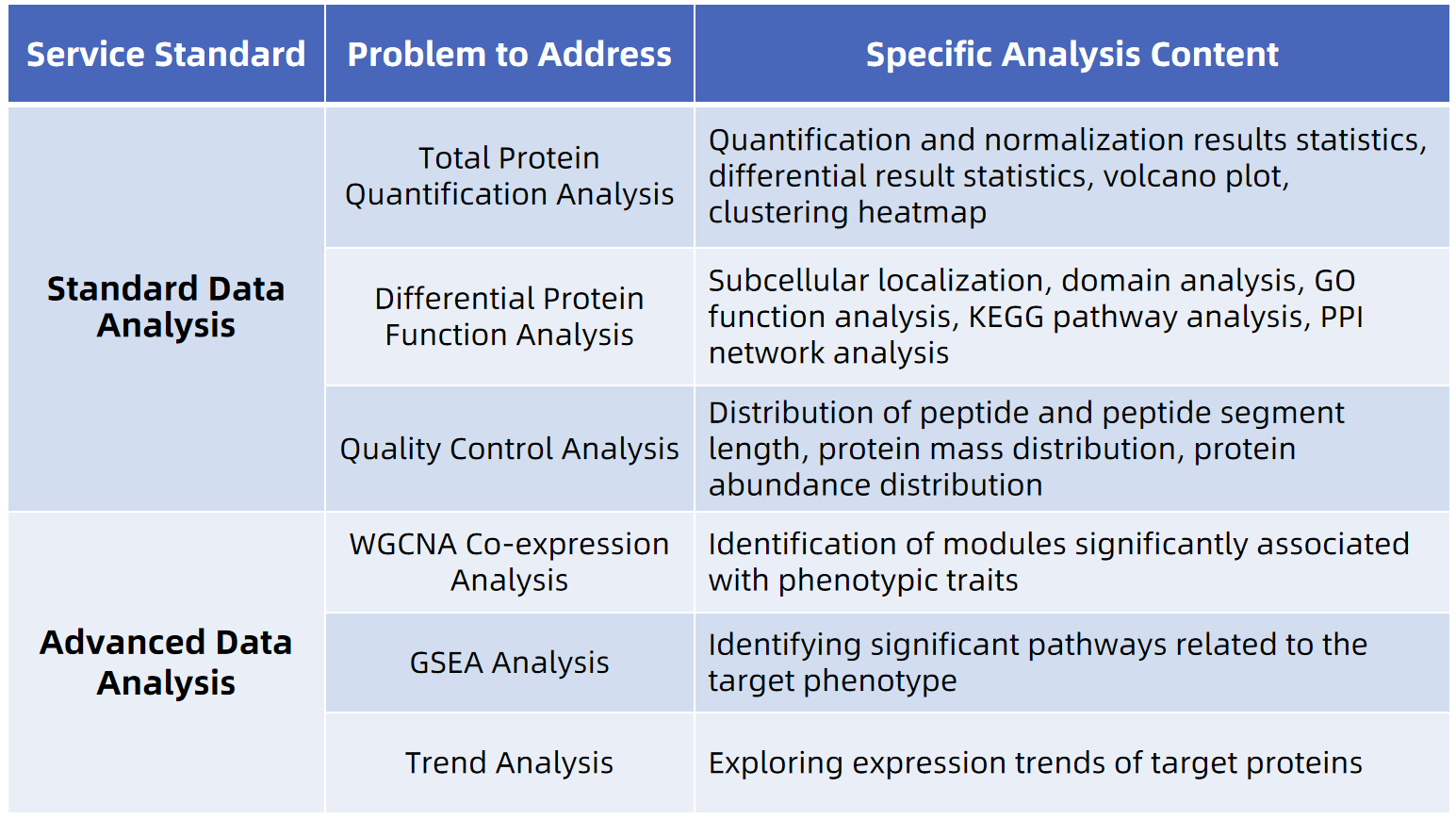
Label-Free Quantitative Proteomics Service, MS Based
- Does not require expensive isotope labeling reagents, resulting in a shorter experimental duration.
- Enables comparative analysis of differential protein expression across different species.
- Overcomes sample number limitations inherent to labeled quantification techniques, offering greater flexibility and convenience.
- Suitable for Analyzing Various Types of Samples, Including Tissue, Cell, Blood, etc.
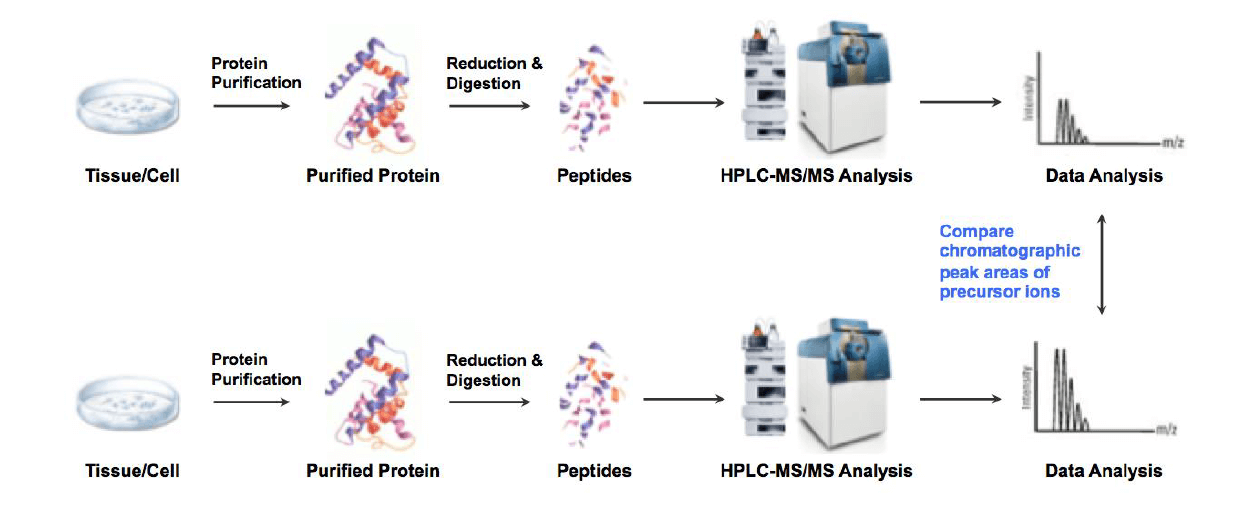
Label-free analysis is a powerful technique for identifying and quantifying relative changes in complex protein samples. In this method, samples are run individually and identified by MS/MS information. Relative quantitation of different samples is then analyzed by aligning chromatographic peak areas of precursor ions between various runs. The most distinctive advantage is that it quantifies proteins without any use of labels, and can analyze unlimited number of samples. Since Label-free is relatively simple and easy compared to other proteomics techniques, it has been widely used for proteomics study and biomarker discovery.
MtoZ Biolabs is proud to offer Label-Free Quantitative Proteomics Service with faster simpler results. We can also perform MS3 analysis to ensure higher analytical accuracy of qualification and quantitation.
Analysis Workflow
Service Advantages
Sample Submission Requirements
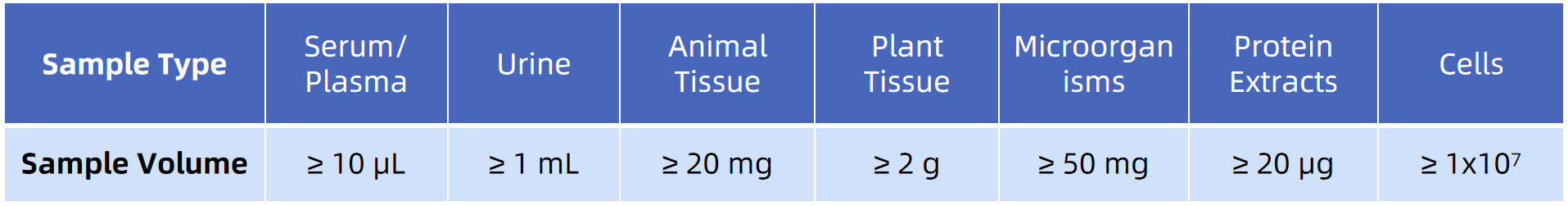
Note: For Co-IP protein samples, only 2-5 ug is required. During sample elution, detergents (such as SDS) and high oxygen concentration should be avoided. Alternatively, you can send the sample beads to us directly.
Bioinformatics Analysis
Deliverables
1. Experiment Procedures
2. Parameters of Liquid Chromatography and Mass Spectrometer
3. MS Raw Data Files
4. Peptide Identifications and Intensities
5. Protein Identifications and Intensities
6. Bioinformatics Analysis
Related Services
Quantitative Proteomics
Protein Analysis
PTM Analysis
How to order?