De Novo Antibody Sequencing Service
Monoclonal antibody plays a key role in the development of biopharmaceuticals and diagnostic kits. The primary structure of the antibody, especially the amino acid sequence of the CDR region, is the core of antibody's biological function. Accurate and rapid analysis of its complete sequence is of great importance for characterization of antibody products. Analysis of the primary antibody sequence is the basis for antibody development and humanization for developing antibody-based drugs. However, due to the variation and modification of antibody products, the sequence information of most antibodies is often not included in the available databases. De novo sequencing technology uses advanced data processing algorithm for sequence analysis, has a superior advantage versus database dependent traditional methods. Antibody sequence can be accurately analyzed using the de novo sequencing method, with no need for database searching.
Based on the world's most advanced mass spectrometry instrument, Orbitrap Fusion Lumos, and combined with rich experience in bioinformatics analysis, MtoZ Biolabs has established a new generation of antibody de novo sequencing platform to achieve accurate analysis of the primary antibody structure. This platform is suitable for sequence analysis of different subtype antibodies, such as IgG and IgM, and different types of antibodies, including fluorescently conjugated antibodies, immobilized antibodies, and antibodies from different species.
Analysis Workflow
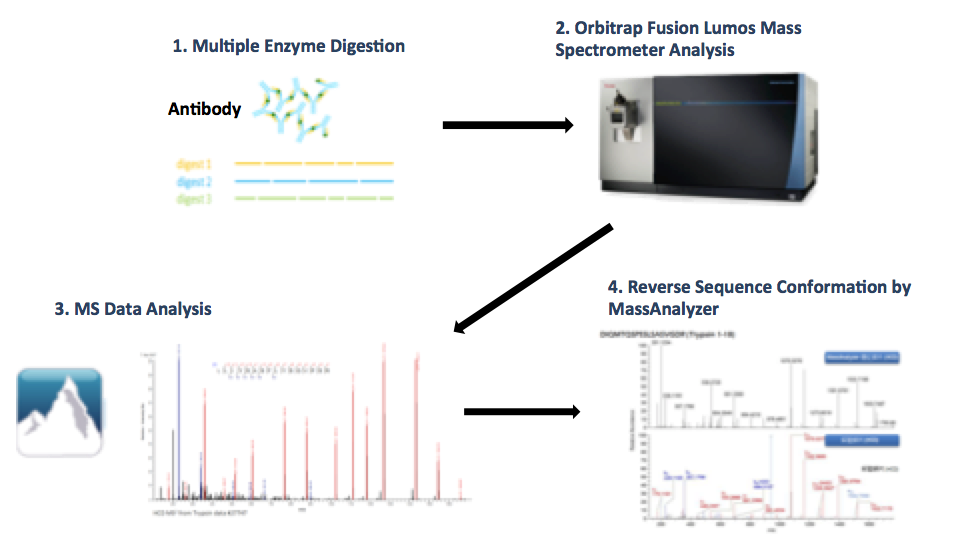
Figure 1
Service Advantages
1. Full Antibody Sequence Coverage
To achieve 100% antibody sequence determination, MtoZ Biolabs selects 5 different proteases for enzyme digestion. 100% full sequence splicing of light and heavy chains of antibody molecules will be achieved by using complementarity between different cleaved peptide segments. The CDR sequence of the antibody light chain is required to be identified at least 10 times during sequence analysis in order to fully ensure the accuracy of the sequencing results (as shown in the figure below).
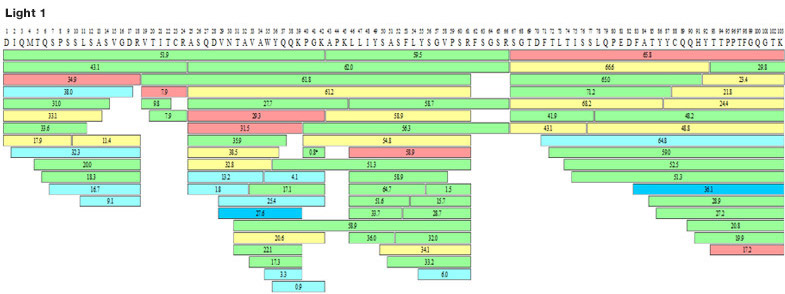
Figure 2. Full Antibody Sequence Coverage
2. Advanced Data Processing Algorithm
Based on the literature published in Nature Biotechnology and professional proteomics magazine JPR, and combined with the professional PEAKS Mass Spectrometry Software, MtoZ Biolabs has developed our own mass spectrometry raw data processing method. This method enables accurate identification of antibody sequences without missing any valid data information.
3. High Precision and Accuracy
MtoZ Biolabs uses Orbitrap Fusion Lumos mass spectrometer with the highest resolution and sensitivity for antibody sequencing. The MS scanning speed of Orbitrap Fusion Lumos has been significantly improved for detecting more peptides, and increases the credibility of the results. Increased MS/MS scanning speed enables detection of more collision-induced fragments (shown below) and accurate determination of leucine and isoleucine can be achieved by analyzing the characteristic peaks in the MS/MS spectrum.
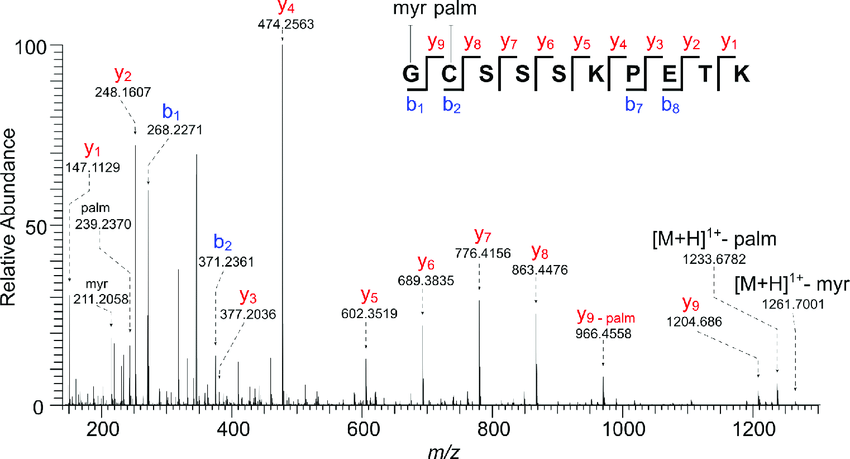
Figure 3. HCD Collision-Induced MS/MS Spectrum
4. Sequencing Multiple Types of Antibodies
Our well-established technology can effectively analyze antibody with modifications of small groups, such as FITC, Biotin, and Alexa. Although large protein modification groups may affect the accuracy of sequencing, the impact can be minimized if the concentration of protein sample is high enough.
5. Sequence Prediction and Strict Sequence Verification
We will first analyze the determined sequence to identify the CDR region of the antibody. At the same time, we establish a new database with the determined sequence, and verify the antibody sequence results using the Sequence Confirmation mode. Only the exact match results will be provided to customers.
Deliverables
1. Experiment Procedures
2. Parameters of Liquid Chromatography and Mass Spectrometer
3. MS Raw Data Files
4. Peptide Identifications and Intensities
5. Antibody Sequencing Results
Related Services
Protein Sequencing
Protein Full-Length Sequencing
Protein Analysis
PTM Analysis
How to order?







