Protein Sequencing Service by Edman Degradation
Edman sequencing method analyzes the protein N-terminal sequence in sequential Edman reactions. While mass spectrometry technology has been commonly used for protein analysis, Edman sequencing is still a powerful and irreplaceable method for protein N-terminal sequencing. As a well-established sequencing method, Edman sequencing provides more accurate protein sequence data, compared to MS. MtoZ Biolabs is aimed at providing reliable protein analysis service. Our protein/peptide N-terminal sequence analysis is performed by SHIMADZU PPSQ-33A Analyzer, equipped with the newest HPLC analysis system.
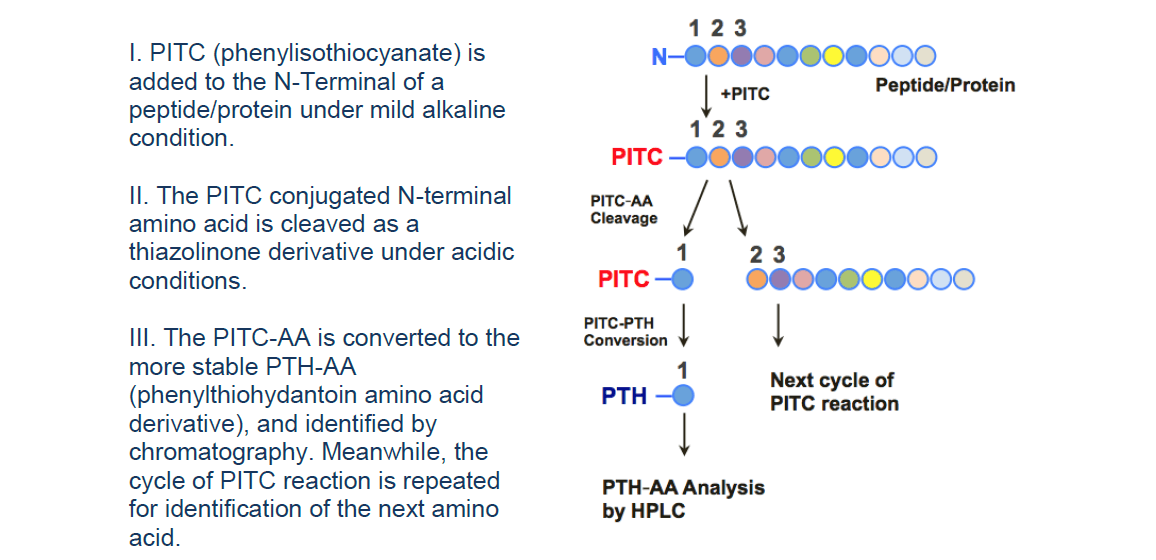
Technical Platforms

Service Advantages
1. High Sensitivity in Detecting PTH-AA at Low Picomole Level
2. Analyze N-Terminal 30 Amino Acids
3. Newest HPLC Analysis System, with UV_VIS SPD-20A Detector
4. Accurately Discriminate Amino Acids of High Similarity, i.e. I/L, and Q/K
Applications
1. Determination of N-Terminal Sequence of Proteins/Peptides/Antibodies/Vaccines
2. Verification of the Correct Translation of Recombinant Proteins
3. Verification of the Sequence of Synthesized Peptides
Sample Submission Requirements
1. Electroblotted Samples: Proteins separated by SDS-PAGE should be blotted onto PVDF membrane. Nitrocellulose is not recommended. PVDF membrane can be stained with Coomassie blue or Poncaue red (Sliver stain is not recommended), followed by washing with ultra-pure water. The washing steps must be repeated several times, when glycine-buffer is used.
2. Liquid Samples: 1-10 ug sample amount, with >90% purity. Avoid using Tris, glycine, guanidine, glycerol, sucrose, ethanolamine, SDS, Triton, X-100, Tween, and ammonium sulfate in the buffer.
*Note: All reagents/solvent used must be of the highest purity to reduce contaminating substances. Samples should be handled with extreme caution and always in clean condition. Any source that may introduce contaminating proteins should be eliminated. Customers are welcome to contact us for detailed sample requirements before sending your samples.
Deliverables
1. Experiment Procedures
2. Parameters and Settings of Edman Degradation Sequencing System
3. PTH-AA HPLC Raw Data Files
4. Protein N-Terminal Amino Acid Sequence
Related Services
Protein Sequencing
Protein Full-Length Sequencing
Protein Analysis
PTM Analysis
How to order?







