De Novo Protein Sequencing Service
- Monoclonal antibodies (mAbs)
- Fab/Fc fragments
- Bispecific antibodies (bsAbs)
- Multispecific antibodies
- Recombinant proteins
- Peptides
- Fluorescently labeled antibodies
- Cross-linked antibodies
- Sample Amount: At least 100 µg.
- Purity Level: A minimum of 90%.
- Sequencing Quantity: At least 20 µg must be available for successful sequencing.
De Novo protein sequencing service is a specialized mass spectrometry-based service that determines the primary structure of proteins by directly analyzing their amino acid sequences without the need for reference genomes or known sequence databases. This service is particularly valuable for studying novel proteins, proteomes of non-model organisms, or identifying unknown or modified proteins in complex samples.
Protein sequences are critical information for study of protein function, as the amino acid sequence determines the advanced protein structures. Generally, there are two methods for protein sequence analysis, database search and de novo sequencing. Database search method identifies proteins by measuring peptide mass, and comparing to available protein databases. However, this method is unsuitable for analyzing antibodies, peptides, and proteins, of which the sequence information is either inaccessible, or not included in any databases. For these cases, de novo sequencing is the only way to decipher the protein sequence.
Service at MtoZ Biolabs
MtoZ Biolabs offers De Novo protein sequencing service utilizing the state-of-the-art Orbitrap Fusion Lumos mass spectrometer, combined with our expertise in bioinformatics analysis and reverse sequence confirmation to ensure high-quality sequence analysis. Our De Novo protein sequencing service is also well-suited for analyzing multiple kinds of antibodies, including IgM and IgG antibody subtypes, and antibodies modified with FITC, biotin, and Alexa, etc., for a wide range of applications:
1. Sequence Analysis of Protein/Peptide, of which Databases are Unavailable
2. PTMs Analysis of Protein/Peptide, of which Databases are Unavailable
3. Sequence Analysis of CDR Region or Full-Length of Antibodies
Analysis Workflow
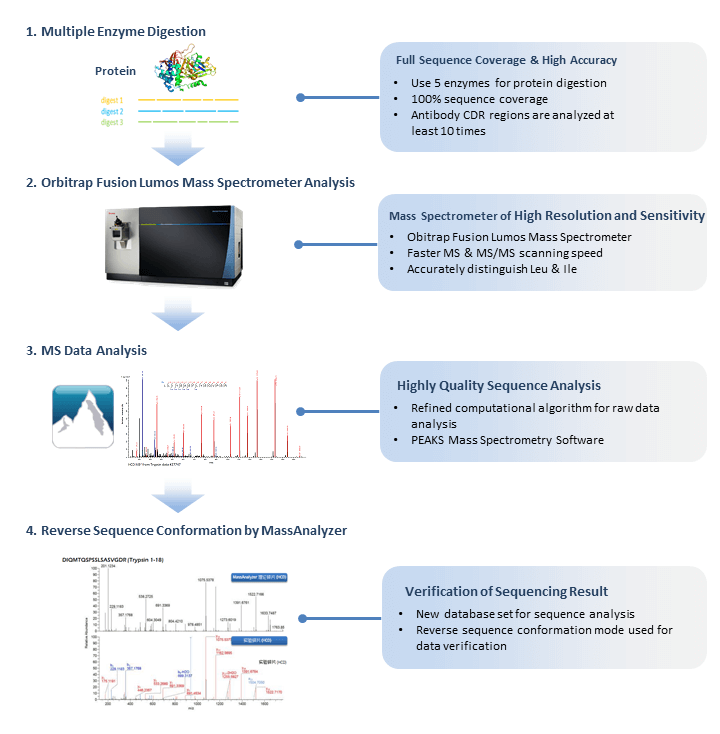
Advantages of De Novo Sequencing Technology
1. No Dependence on Reference Sequences
De Novo protein sequencing does not rely on genomes or sequence databases, enabling de novo determination of amino acid sequences for unknown proteins. This makes it particularly suitable for studying non-model organisms or novel proteins.
2. High Sensitivity and Precision
Utilizing high-resolution mass spectrometers such as Orbitrap and FTICR-MS, the technique can accurately detect low-abundance proteins and complex samples, resolving peptide amino acid sequences with exceptional reliability.
3. Comprehensive Analysis of Post-Translational Modifications (PTMs)
Precisely identifies protein PTMs such as phosphorylation and glycosylation, including their types, sites, and functional impacts, providing critical insights for advanced research.
4. Broad Applicability
Supports analysis of purified proteins, complex mixtures, and unknown proteins in samples, making it ideal for antibody sequencing, protein function analysis, and biopharmaceutical development.
Case Study
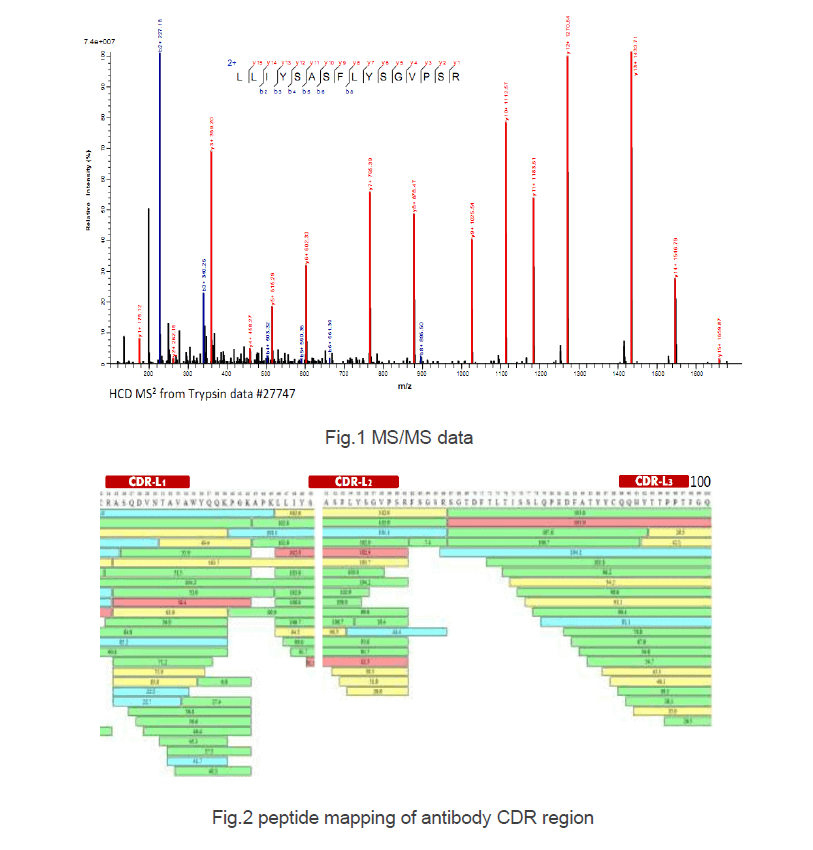
In this study, the full-length sequence of a monoclonal antibody is analyzed by our de novo sequencing system. MS/MS data in Fig. 1 indicated that complete fragmentation of peptide precursors and L/I residues can be easily discriminated after second collision. We used four enzymes (indicated by different color in Fig. 2) for protein digestion. Peptide mapping results showed complete sequence coverage with CDR being identified by at least 10 times, reaffirming accuracy of our sequence analysis.
Why Choose MtoZ Biolabs?
1. Cutting-Edge Mass Spectrometry Platform
MtoZ Biolabs is equipped with industry-leading mass spectrometry instruments, including Thermo Fisher Orbitrap series, FTICR-MS, and Nano-LC systems, ensuring high resolution and precision for De Novo protein sequencing.
2.Expert Technical Team
Our team of experienced PhDs and technical experts has extensive knowledge in proteomics and bioinformatics, ensuring the scientific rigor and reliability of every project.
3.Comprehensive Service Solutions
We offer end-to-end services, including sample preparation, mass spectrometry analysis, and data interpretation, covering unknown protein identification, antibody sequencing, and post-translational modification (PTM) analysis to meet diverse research needs.
4. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
5. High-Data-Quality
Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all De Novo protein sequencing service data, providing clients with a comprehensive data report.
Deliverables
1. Experiment Procedures
2. Parameters of Liquid Chromatography and Mass Spectrometer
3. MS Raw Data Files
4. Peptide Identifications, Intensity, and Peptide Mapping
5. Protein Sequence and PTMs Data
FAQs
Q1. What types of samples are suitable for De Novo protein sequencing?
A: MtoZ Biolabs’ De Novo protein sequencing service supports a wide range of sample types, regardless of protein size or length. It is highly versatile and applicable to the following sample categories, including but not limited to:
This technology is flexible enough to deliver precise sequencing results regardless of the size or complexity of the protein sample.
Q2. What are the sample requirements for De Novo protein sequencing?
A: To ensure the accuracy of the results, the sample should meet the following criteria:
It is important to note that sample purity significantly impacts sequencing outcomes. Contaminants such as antibody proteins, homologous proteins, or bovine serum albumin (BSA) may interfere with the sequencing process, reducing the reliability of the results.
For further questions, feel free to contact MtoZ Biolabs, and our experts will provide professional answers and support tailored to your needs!
Related Services
Protein Sequencing
Protein Full-Length Sequencing
Protein Analysis
PTM Analysis
How to order?







