Ubiquitination Site Identification Service
- Protein Degradation Studies: Identify ubiquitination sites involved in proteasome-mediated degradation pathways.
- Cell Signaling Analysis: Explore how ubiquitin modifications regulate key signaling cascades.
- Disease Mechanism Research: Uncover dysregulated ubiquitination patterns in cancer, neurodegeneration, and immune disorders.
- Drug Target Validation: Evaluate ubiquitination status of candidate proteins in response to therapeutic compounds.
- E3 Ligase Substrate Discovery: Map substrates of specific E3 ubiquitin ligases to reveal regulatory networks.
- Storage and Shipping: Samples should be stored at -80℃ and shipped with dry ice.
Protein ubiquitination is a reversible post-translational modification in which ubiquitin molecules are covalently attached to substrate proteins, thereby modulating their stability, localization, activity, and interactions. MtoZ Biolabs, a leading provider of advanced proteomics services, offers precise and comprehensive Ubiquitination Site Identification Service to support the discovery and functional analysis of ubiquitin-mediated signaling pathways across diverse research contexts.
Ubiquitination involves the attachment of ubiquitin, a highly conserved 76-amino acid protein, to lysine residues of substrate proteins through an enzymatic cascade that includes E1 activating enzymes, E2 conjugating enzymes, and E3 ligases. This sequential process results in either monoubiquitination or the assembly of polyubiquitin chains with various linkage types such as K48, K63, or linear chains. Each linkage type conveys distinct biological signals. Ubiquitin modification regulates numerous cellular processes by directing proteins for proteasomal degradation, altering subcellular localization, modulating enzymatic activity, or influencing protein–protein interactions. Beyond targeting proteins for proteasomal degradation, ubiquitination also plays crucial roles in regulating protein turnover, DNA damage response, autophagy, cell cycle progression, endocytosis, and innate immunity.
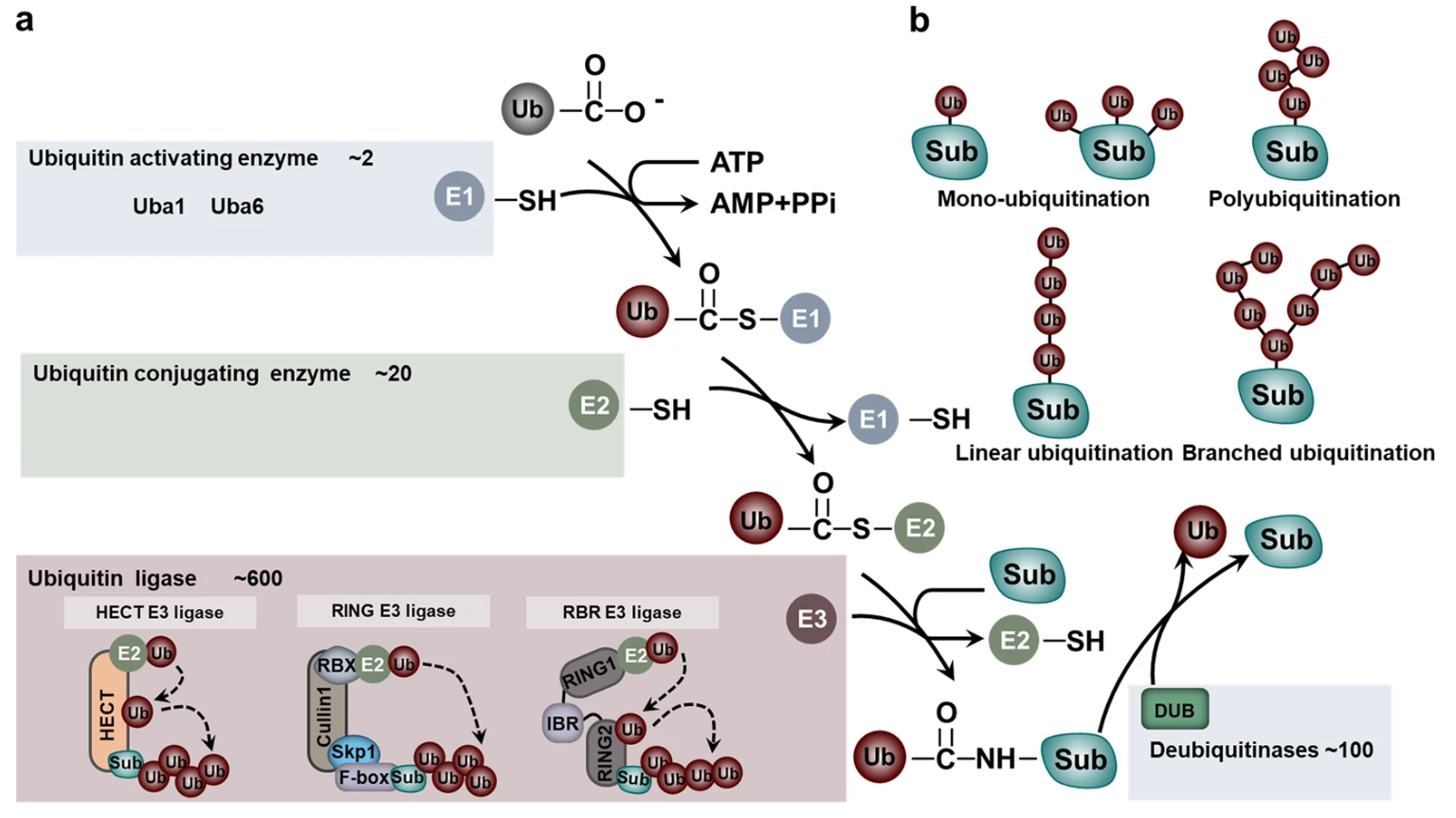
Figure 1. The Components and Processes of the Ubiquitin Proteasome System
Service at MtoZ Biolabs
The dynamic and transient nature of ubiquitination, combined with its low stoichiometry in vivo, poses a significant analytical challenge, particularly when aiming for precise site-level resolution in complex biological matrices. To meet these challenges, MtoZ Biolabs provides a high-performance Ubiquitination Site Identification Service based on high-resolution LC-MS/MS combined with advanced peptide enrichment technologies. Our service enables sensitive and site-specific identification of ubiquitinated lysine residues, quantitative assessment of modification dynamics, and comprehensive coverage of ubiquitin signaling networks. With customizable workflows, rigorous quality control, and expert bioinformatics analysis, our ubiquitination site identification platform is ideal for studies involving proteostasis, drug mechanism-of-action, E3 ligase substrate screening, and more. In addition to ubiquitination, we also offer specialized proteomics solutions for other post-translational modifications, including phosphorylation, acetylation, glycosylation, and more, enabling integrated analyses of PTM crosstalk and functional regulation.
Analysis Workflow
Our analytical workflow for ubiquitination site identification includes protein extraction, enzymatic digestion, enrichment of ubiquitinated peptides, high-resolution LC-MS/MS analysis, data processing, and comprehensive bioinformatics analysis.

Figure 2. Workflow for Ubiquitination Site Identification Service
Service Advantages
1. End-to-End Service
From sample preparation to data interpretation, MtoZ Biolabs provides a streamlined, one-stop solution to accelerate ubiquitin-related research.
2. Experienced Team
Backed by a team of proteomics experts and years of experience in PTM analysis, we deliver high-quality, reproducible results trusted by academic and industry clients.
3. Comprehensive Coverage
Our optimized workflows enable detection of both mono- and polyubiquitination across diverse linkage types, offering a complete view of ubiquitin signaling networks.
4. Customizable Analysis Pipelines
We offer flexible experimental designs and data processing strategies tailored to specific research goals, from global profiling to targeted validation.
Applications
Sample Submission Suggestions

*Note: If you have specific requirements or need guidance on sample preparation, please do not hesitate to contact us.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Our Ubiquitination Site Identification Service is designed to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Contact us today to discuss your project and request a custom proposal!
Related Services
Post-Translational Modifications Proteomics Service
PRM-based Post-translational Modification Site Analysis Service
Post-Translational Modification Site Motif Analysis Service
How to order?







