Phosphorylation Site Identification Service
Phosphorylation site identification is a proteomics analysis service focused on detecting and locating phosphorylation sites on proteins. Phosphorylation is one of the most common post-translational modifications, referring to the covalent attachment of a phosphate group to serine (Ser), threonine (Thr), or tyrosine (Tyr) residues. It plays a central role in regulating biological processes such as signal transduction, cell cycle progression, and metabolic responses. This service typically integrates efficient phosphopeptide enrichment techniques (such as TiO₂ or IMAC) with high-resolution mass spectrometry platforms to identify or predict phosphorylation sites within specific proteins or across the proteome, and provides corresponding peptide sequences and spectral information.
The phosphorylation site identification service is widely applied in areas such as signaling pathway research, kinase target screening, drug mechanism studies, biomarker discovery, and the investigation of tumor and immune regulation. It is particularly valuable for uncovering key phosphorylation events involved in dynamic regulatory processes and helps elucidate molecular regulatory networks under physiological and pathological conditions.
Services at MtoZ Biolabs
1. Target Protein Phosphorylation Site Identification
MtoZ Biolabs provides Target Protein Phosphorylation Site Identification, designed to accurately detect and characterize phosphorylation sites on specific proteins of interest. By integrating phospho-specific antibody enrichment with high-resolution LC-MS/MS analysis, we precisely localize modified residues and quantify phosphorylation levels. This service is ideal for validating signaling-related proteins, studying regulatory mechanisms, and confirming phosphorylation-dependent functional changes in targeted biological systems.
2. Phosphoproteomics Site Identification
MtoZ Biolabs' Phosphoproteomics Site Identification enables comprehensive, proteome-wide mapping of phosphorylation sites across diverse biological samples. Utilizing advanced phosphopeptide enrichment techniques such as TiO₂, IMAC, or Fe³⁺-IMAC combined with high-resolution mass spectrometry and multi-mode fragmentation (HCD, ETD, EThcD), we systematically identify and quantify thousands of phosphorylation sites. This large-scale approach reveals signaling pathway activation, dynamic modification networks, and potential biomarkers, providing researchers with deep insights into cellular regulation and disease-related phosphorylation events.
Analysis Workflow
1. Protein Extraction and Digestion
Proteins are extracted from cells, tissues, or purified samples. After reduction and alkylation, trypsin digestion is performed to generate peptides while preserving phosphorylation information.
2. Phosphopeptide Enrichment
TiO₂ or IMAC-based enrichment methods are employed to selectively isolate phosphopeptides, improving the detection sensitivity of low-abundance phosphorylation sites.
3. LC-MS/MS Analysis
High-resolution mass spectrometry platforms (e.g., Orbitrap) are used to perform liquid chromatography–tandem mass spectrometry on enriched peptides, generating high-quality spectral data.
4. Data Analysis and Site Localization
Modification sites are identified through database searching and specialized phosphorylation algorithms (such as Ascore or PhosphoRS), providing annotated phosphopeptides, site positions, and confidence scores.
Sample Submission Suggestions
1. Sample Types
Supports a wide range of biological materials, including cells, tissues, serum/plasma, and purified proteins. It is recommended to use samples with high phosphorylation activity or known active signaling pathways to improve site detection efficiency.
2. Buffer Requirements
Avoid using buffers containing SDS, glycerol, high salt, or phosphate-based components that may interfere with mass spectrometry analysis. Use MS-compatible buffers for optimal results.
3. Sample Transport
Samples should be stored at –80°C and shipped on dry ice to maintain phosphorylation stability and prevent degradation from freeze-thaw cycles. Sample handling guidance is available upon request.
Service Advantages
1. High-Resolution Site Localization
Utilizes high-end mass spectrometry platforms such as Orbitrap, combined with phosphopeptide-specific enrichment strategies, to precisely identify and localize phosphorylation sites on Ser, Thr, and Tyr residues.
2. Strong Detection of Low-Abundance Modifications
Employs optimized TiO₂ or IMAC enrichment methods to significantly enhance the detection of low-abundance phosphopeptides, making it suitable for complex biological samples.
3. Rigorous and Reliable Data Analysis
Incorporates multiple scoring algorithms to evaluate site confidence, improving the accuracy and reproducibility of phosphorylation site identification.
4. End-to-End Customized Service
Covers the complete workflow from sample preparation, phosphopeptide enrichment, and mass spectrometry detection to data processing and interpretation, providing flexible and customized solutions tailored to diverse research objectives.
Applications
1. Signal Pathway Mechanism Research
The phosphorylation site identification service is used to analyze kinase-substrate interactions, phosphorylation cascades, and regulatory signaling networks, contributing to a deeper understanding of cellular response mechanisms.
2. Drug Mechanism Validation
By comparing phosphorylation levels before and after treatment, the service helps evaluate the impact of candidate drugs on key protein phosphorylation, supporting pharmacodynamic studies and target validation.
3. Cancer and Disease Research
Applicable for identifying aberrant phosphorylation events associated with diseases such as cancer, neurodegenerative disorders, and immune dysfunctions, facilitating disease mechanism exploration and target discovery.
4. Kinase Target Discovery
This service aids in identifying kinase-regulated sites and substrate proteins, providing valuable insights for kinase screening, functional validation, and inhibitor development.
Case Study
1. Identification and Functional Analysis of SOX10 Phosphorylation Sites in Melanoma
This study aimed to investigate the phosphorylation modifications of the SOX10 protein in melanoma and their functional implications. Using the human melanoma cell line 501mel as a model, several phosphorylation sites on SOX10 were identified through co-immunoprecipitation combined with LC-MS/MS, including S24, S45, and T240, which are located in predicted MAPK/CDK recognition motifs. Phosphorylation-deficient mutants were constructed for functional validation. The mutations did not alter SOX10 subcellular localization but showed cell-specific effects on the transcriptional activation of the MITF promoter. Protein stability assays revealed that the S24A and T240A mutants had significantly shorter half-lives compared to the wild-type protein. These findings suggest that SOX10 phosphorylation regulates its stability and transcriptional activity and plays a crucial role in melanoma development, indicating its potential as a therapeutic intervention target.
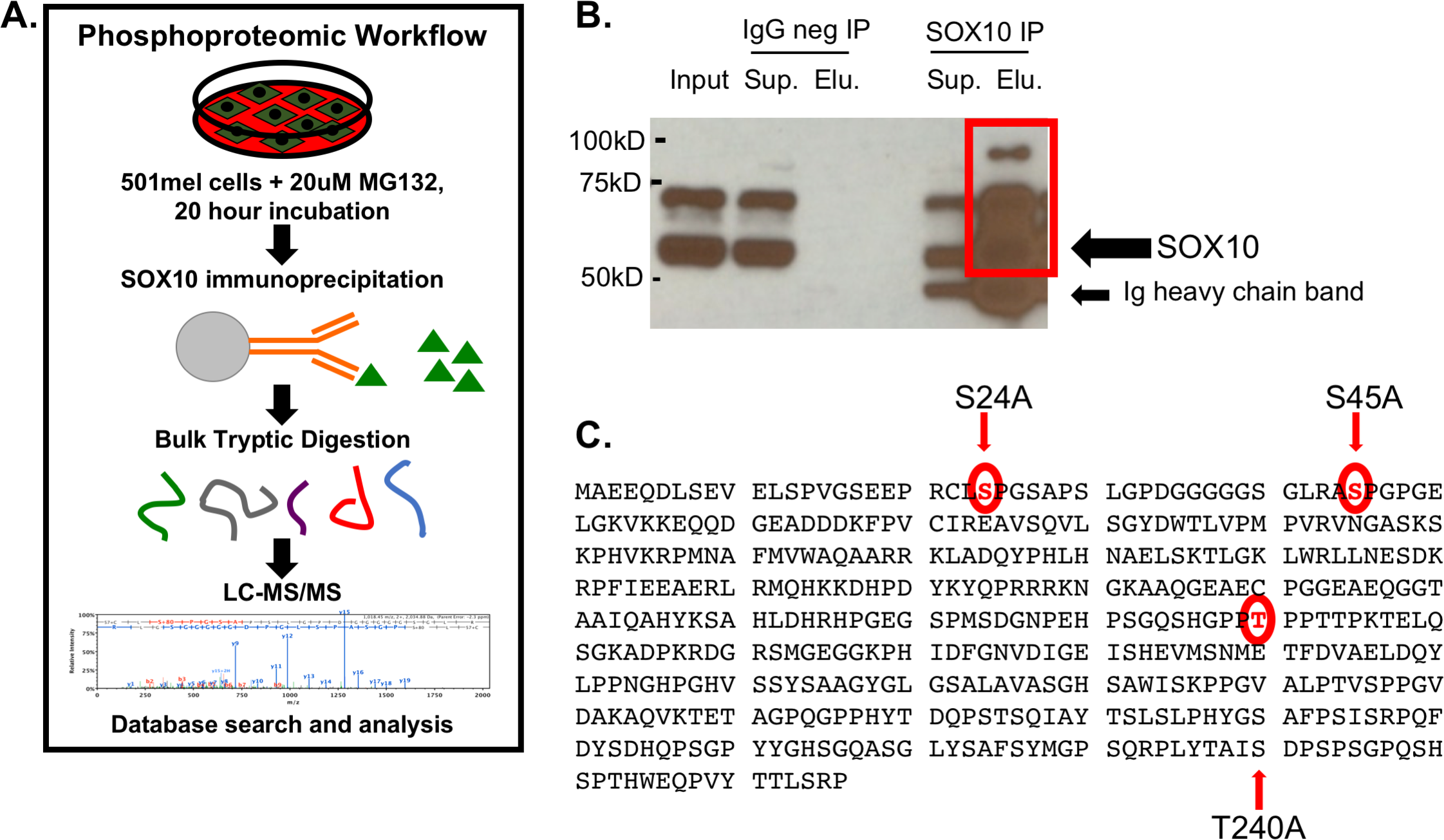
Cronin, J C. et al. PLOS ONE, 2018.
Figure 1. Mass Spectrometry Analysis Identifies SOX10 Phosphorylation Sites.
2. Identification of Phosphorylation Sites on β1-adrenergic Receptor in the Mouse Heart
This study aimed to identify the phosphorylation sites on the β1-adrenergic receptor (Adrb1) in mouse heart tissue to better understand its role in cardiac function regulation. Using mouse heart samples, researchers employed immunoaffinity purification combined with nano-scale liquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) and identified phosphorylation at Ser274 and Ser280 within the third intracellular loop, as well as Ser412, Ser417, Ser450, Ser451, and Ser462 at the C-terminal region of Adrb1. Further analysis revealed that stimulation with an agonist increased phosphorylation at Ser274, Ser280, and Ser462. This study represents the first in vivo systematic identification of Adrb1 phosphorylation sites and provides new insights into the regulatory mechanisms of β1-adrenergic receptor function in the heart.
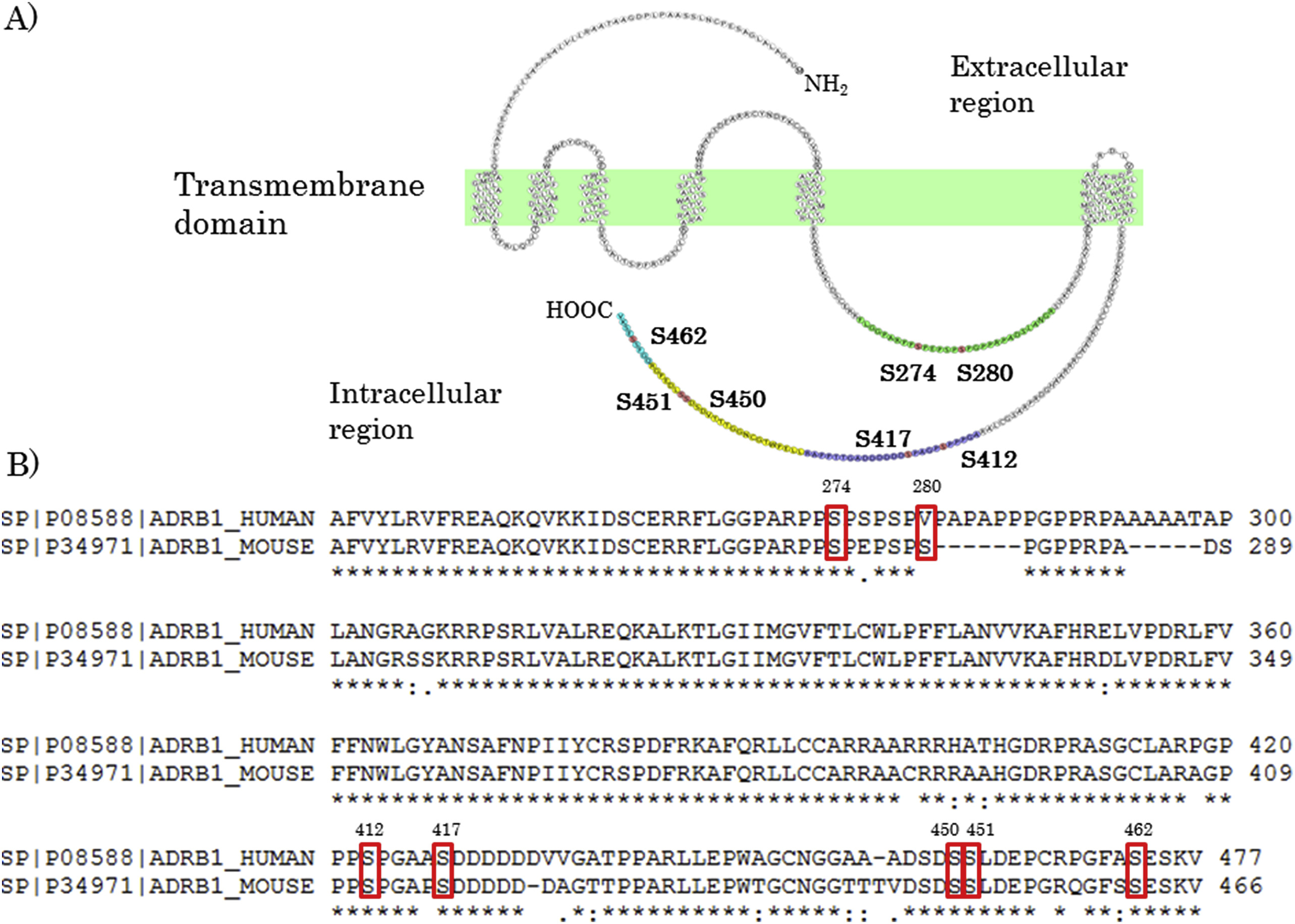
Hayashi, K. et al. Biochemical and Biophysical Research Communications, 2017.
Figure 2. Phosphorylated Residues Were Detected at the Third Intracellular Loop and C-Terminal of Adrb1.
FAQ
Q1: What Types of Phosphorylation Modifications Can this Service Identify?
A1: This service primarily identifies three common types of phosphorylation sites: serine (Ser), threonine (Thr), and tyrosine (Tyr). It is applicable to various sample types, including cells, tissues, and purified proteins.
Q2: Can Low-Abundance Phosphorylation Sites Be Detected?
A2: Yes. We utilize highly efficient enrichment techniques (such as TiO₂ and IMAC) combined with high-sensitivity mass spectrometry platforms (such as Orbitrap), which significantly enhance the detection of low-abundance modified peptides.
Q3: Can Multiple Sample Groups Be Compared?
A3: Yes. Label-free or TMT-based quantitative strategies can be used to compare phosphorylation level changes across different treatment groups, making this service suitable for signaling pathway studies and drug mechanism exploration.
How to order?







