tRNA Modification LC-MS Analysis Service
- Analysis of modification profiles in tumors, mitochondria, and neurodegenerative diseases
- Mechanistic studies in modification enzyme knockout (KO/KD) models
- Construction of translation regulation profiles under different cellular conditions
- RNA modification biomarker discovery and personalized diagnostic exploration
- Optimization of modification control in synthetic biology and RNA engineering
tRNA plays an essential role in protein translation by decoding mRNA codons into corresponding amino acids. Its functionality is critically dependent on a variety of post-translational modifications (PTMs) that ensure proper structural conformation, protect it from degradation, and enhance translation efficiency and accuracy. tRNA modifications are also involved in cellular stress responses and the regulation of gene expression.
As the most modified RNA species, tRNA modifications are found across functional domains such as the D-loop, T-loop, and anticodon loop. These modifications include methylation (e.g., m⁵C, m¹A), pseudouridylation (Ψ), thiolation (s²U), and inosine (I), among others. Dysregulated tRNA modifications have been associated with several diseases, including cancer, neurodegenerative disorders, metabolic diseases, and mitochondrial dysfunction.
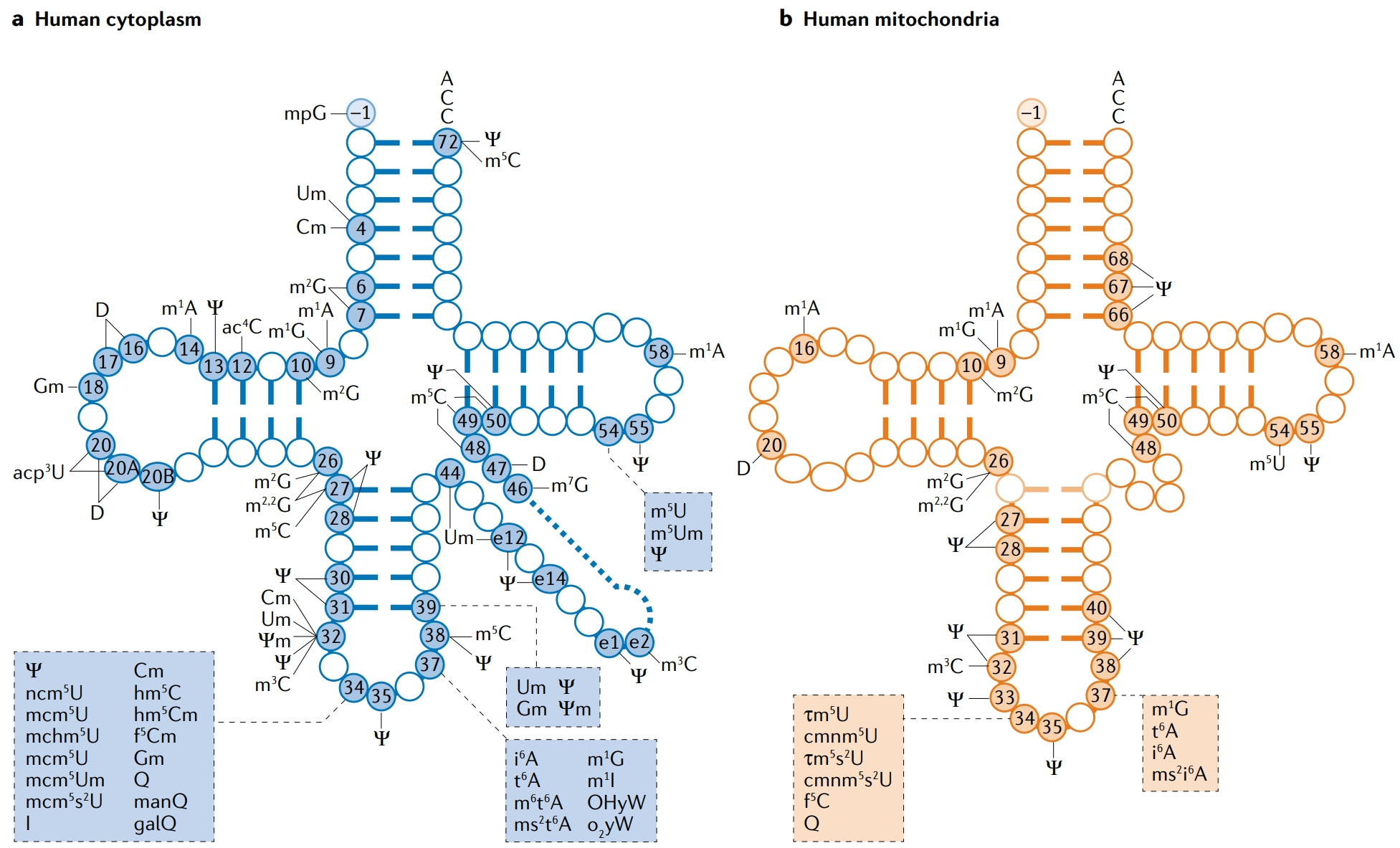
Suzuki, T. et al. Nat. Rev. Mol. Cell Biol. 2021.
Figure 1. Typical Distribution of tRNA Modifications in Human Cytoplasm and Mitochondria
To support systematic exploration of tRNA modification research, MtoZ Biolabs offers tRNA Modification LC-MS Analysis Service which uses advanced mass spectrometry and enrichment technologies to identify and compare modification pattern variations across different samples, facilitating functional research and the development of translational applications.
Services at MtoZ Biolabs
1. Targeted tRNA Modification Analysis
MtoZ Biolabs provides Targeted tRNA Modification Analysis for quantitative detection of specific tRNA modifications designated by clients. Using high-resolution LC-MS and optimized chromatographic methods, we accurately identify and quantify defined modification sites with high precision.
2. Global tRNA Modification Profiling
For large-scale analysis, MtoZ Biolabs offers Global tRNA Modification Profiling to comprehensively assess diverse tRNA modifications across samples. This full-spectrum LC-MS workflow enables systematic mapping and quantitative comparison of modification patterns for discovery-driven studies.
Our tRNA Modification LC-MS Analysis Service provides a quantitative detection system covering over 50 nucleoside modifications. The detectable modification types include:
· Methylation Modifications: Such as m⁵C, m¹A, m⁷G, m²²G, involved in tRNA structural stability and translation regulation.
· Deamination Modifications: Inosine (I), enhancing codon recognition flexibility.
· Thiolation Modifications: Such as s²U, mcm⁵s²U, which improve thermal stability and translation efficiency.
· Acetylation/Amidation Modifications: Such as ac⁴C, t⁶A, involved in conformation regulation and enzyme recognition.
· Isomerization and Special Functional Modifications: Such as Ψ (Pseudouridine), hm⁵C, and Q (Queuosine), regulating tRNA half-life and translation accuracy.
Technical Principles
tRNA modification LC-MS analysis leverages subtle differences in the structure, mass, and polarity of modified nucleosides. These modifications exhibit distinct retention times in liquid chromatography, and in mass spectrometry, they show characteristic parent and fragment ion spectra. By comparing with reference standards or databases, the chemical structure and abundance of each modification can be precisely identified. For modifications with complex structures or close mass shifts, multi-stage tandem mass spectrometry (MS/MS) and isotope labeling techniques are employed for structural elucidation and relative/absolute quantification.
With this approach, MtoZ Biolabs can accurately identify and perform relative/absolute quantification of dozens of tRNA-modified nucleosides in complex samples, enabling differential analysis across multiple sample sets.
Service Advantages
✅ Advanced Analysis Platform:: Utilize Orbitrap Fusion Lumos, Q Exactive HF, and other high-resolution systems to ensure precise, reliable detection of low-abundance tRNA modifications
✅ Comprehensive Modification Profiling: Identify and quantify over 50 types of tRNA modifications across a variety of functional domains
✅ Parallel Multi-Modification Analysis: Simultaneously analyze multiple modification types, saving time and reducing experimental costs
✅ Tailored Analytical Solutions: Customizable workflows to meet your specific experimental design, sample type, and research objectives
✅ Experienced Technical Team: Expertise in handling complex biological samples, from cell lines and tissues to mitochondrial and RNA engineering samples
Sample Submission Suggestions
High-quality samples are essential for obtaining reliable modification data. To ensure that the results of the tRNA Modification LC-MS Analysis Service is reproducible and biologically relevant, we recommend that clients follow these sample preparation guidelines:
· Sample Types: Cells, tissues, blood, FFPE samples, etc.
· Recommended Amount: Total RNA ≥ 20 µg; purified tRNA ≥ 5 µg.
· Storage Conditions: RNase-free tubes, stored at -80°C, transported on dry ice.
· Preprocessing Support: We offer services including RNA extraction, tRNA purification, and other sample preparations.
We will provide detailed sample submission guidelines and transportation instructions to ensure the integrity of your samples and high-quality analysis results.
Applications
With the comprehensive modification detection capabilities of the tRNA Modification LC-MS Analysis Service, MtoZ Biolabs provides broad support for a range of basic and applied research projects, including disease mechanism analysis, functional validation, and molecular biomarker development.
Deliverables
1. Raw Data Files (available in .raw or .mzML format).
2. Modification Quantification Results (including relative abundance, p-values, and other statistical data).
3. Modification Type Annotation Report (based on the MODOMICS database).
4. Differential Analysis Charts (e.g., bar charts, heatmaps, volcano plots).
5. Customizable Visualization Report (PDF or PPT format, ideal for presentations and reporting).
We offer comprehensive data interpretation and technical support, ensuring that results are ready for direct use in research reports and academic publications.
FAQ
Q1: Can isomeric modifications (e.g., m⁵C vs hm⁵C) be distinguished?
Yes. Using high-resolution mass spectrometry combined with optimized chromatography, we can differentiate closely related isomers, and further increase accuracy using fragmentation ion analysis.
At MtoZ Biolabs, we offer high-quality, reliable technical support for RNA modification research. Our tRNA Modification LC-MS Analysis Service provides an in-depth, customizable solution for exploring the roles of RNA modifications in biological processes, disease research, and drug development.
Contact us today for a tailored proposal and pricing information.
Related Services
5-methyluridine Analysis Service
How to order?







