Isotope Labeling-Based Quantitative Service
Isotope Labeling-Based Quantitative Service utilizes isotope labeling strategies (such as SILAC, TMT/iTRAQ, and Dimethyl Labeling) combined with high-resolution mass spectrometry (MS) analysis to achieve accurate quantification of proteins or peptides. Isotope labeling-based quantification is one of the most widely used protein quantification methods, relying on stable isotope incorporation into peptides or proteins. This results in mass shifts during MS analysis, enabling precise multi-sample quantification. Compared to Label-Free Quantification (LFQ), isotope labeling methods offer higher accuracy, better reproducibility, and are particularly suitable for complex samples, parallel multi-sample analysis, and highly sensitive detection of low-abundance proteins. Leveraging an advanced mass spectrometry platform, MtoZ Biolabs provides Isotope Labeling-Based Quantitative Service to analyze protein expression levels and modification states under different experimental conditions. We offer comprehensive solutions from cell culture to bioinformatics analysis. Contact us to learn more.
Analysis Workflow
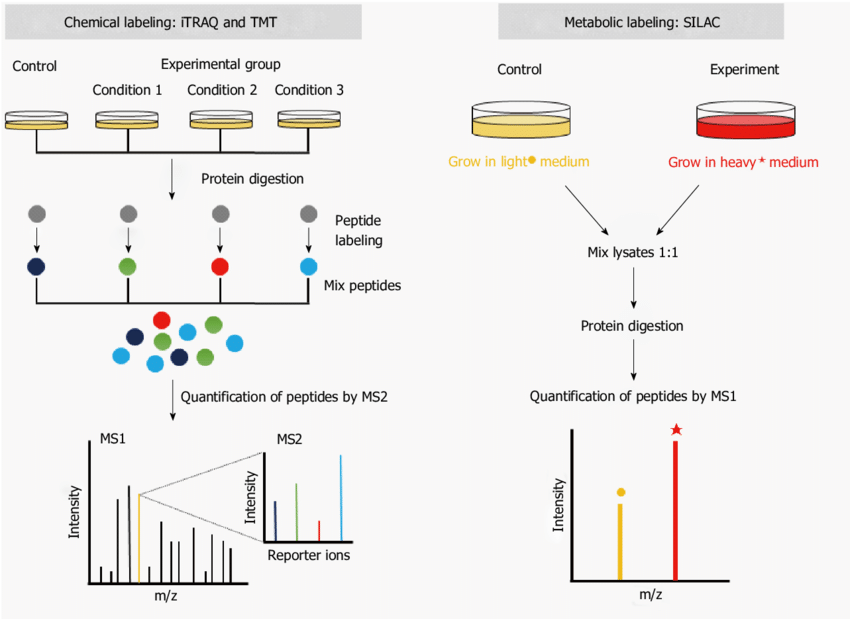
Kang C. et al. World Journal of Gastroenterology. 2016.
1. Sample preparation and isotope labeling
Choose an appropriate isotope labeling strategy according to experimental requirements:
SILAC (Stable Isotope Labeling by Amino acids in Cell culture) requires labeling with light (¹²C, ¹⁴N) and heavy (¹³C, ¹⁵N) amino acids during cell culture, so that the proteins synthesized by the cells carry different stable isotope labels. Then perform protein extraction and enzymatic hydrolysis.
TMT (Tandem Mass Tag) and iTRAQ (Isobaric Tags for Relative and Absolute Quantification) require the introduction of TMT (6-plex, 10-plex, 16-plex) or iTRAQ (4-plex, 8-plex) labels after protein extraction and enzymatic hydrolysis to generate peptides.
2. Mass spectrometry detection
Use Nano-LC combined with high-resolution mass spectrometry to analyze peptides. If post-translational modification (PTM) analysis is required, targeted enrichment of modified peptides is performed prior to MS detection.
3. Data analysis and quantification
Perform data processing and isotope labeling quantitative calculations. Analyze the biological functions of proteins through KEGG, GO enrichment analysis, PPI interaction network, etc.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. MtoZ Biolabs offers isotope labeling-based quantitative service including SILAC, TMT, and iTRAQ, to accommodate diverse research needs.
Applications
Protein Expression Quantification Analysis
Compare protein expression levels under different physiological or pathological conditions, such as disease vs. healthy groups or before and after drug treatment.
Isotope Labeling-Based Quantitative is suitable for biomarker discovery and validation.
Post-Translational Modification (PTM) Quantification
By combining enrichment strategies for modifications such as phosphorylation, acetylation, and ubiquitination, Isotope Labeling-Based Quantitative can be used to study the dynamic changes of specific modifications.
Explore the regulatory mechanisms of cellular signaling pathways.
Drug Mechanism of Action Research
Using Isotope Labeling-Based Quantitative service to assess the impact of drug treatment on protein expression levels in cells or tissues.
Study drug targets and resistance mechanisms.
Clinical Research
Evaluate the specificity and reliability of biomarkers to support clinical diagnosis and precision medicine.
FAQ
Q: How can the appropriate isotope labeling method be chosen?
The choice of an isotope labeling method depends on experimental objectives, sample type, experimental throughput, and data quantitative precision. Common methods and their applicable scenarios are as follows:
SILAC: Suitable for cell culture samples, enabling in vivo labeling to minimize technical errors between samples. It is ideal for protein interaction studies or whole-proteome quantification.
TMT/iTRAQ: Suitable for complex tissues or clinical samples, offering multiplex labeling (up to 16 channels) to increase experimental throughput. However, high-resolution mass spectrometry (e.g., Orbitrap) is required to resolve reporter ion signals.
Dimethyl Labeling: A chemical labeling method that is suitable for large-scale proteomic quantification, offering a lower cost. However, interference from isotope peaks needs to be optimized.
When choosing a method, it is essential to consider sources of experimental error. For example, SILAC may be limited by the conversion of essential amino acids, while TMT/iTRAQ may experience reporter ion suppression effects in low-complexity samples.
Q: How can the efficiency of isotope labeling be assessed?
Assessing isotope labeling efficiency is crucial as it affects both quantification accuracy and experimental reproducibility.
1. SILAC Labeling Efficiency Assessment
Analyze the peak area ratio between light and heavy-labeled peptides (efficiency is considered high if >95%) by MS1 spectrum analysis.
Use an internal control protein, such as GAPDH, to verify that all cells have been sufficiently labeled.
Monitor unlabelled peptides. If there is a high proportion of unlabelled peptides, extend the culture time or optimize the culture medium formula to improve labeling efficiency.
2. TMT/iTRAQ Labeling Efficiency Assessment
After labeling a single standard protein (e.g., BSA), perform LC-MS/MS analysis to check for incomplete labeling of peptides.
Calculate the signal balance of TMT/iTRAQ reporter ions. If there is a systematic bias between channels, the sample mixing ratio or labeling reaction conditions may need optimization.
Precise evaluation of labeling efficiency is essential for ensuring data reliability. It is recommended to combine LC-MS optimization with internal standards for quality control throughout the experiment.
Case Study
This study employed TMT-based quantitative proteomics to investigate the therapeutic effects of Lianling decoction (LLT) on atopic dermatitis (AD) induced by Staphylococcus aureus infection. The results showed that LLT significantly reduced skin inflammation scores, epidermal thickening, and mast cell infiltration in AD model mice, while also lowering serum levels of IgE, IL-4, and IL-6. Proteomics analysis revealed that LLT may exert its therapeutic effect by modulating the expression of proteins related to immune responses and inflammation.
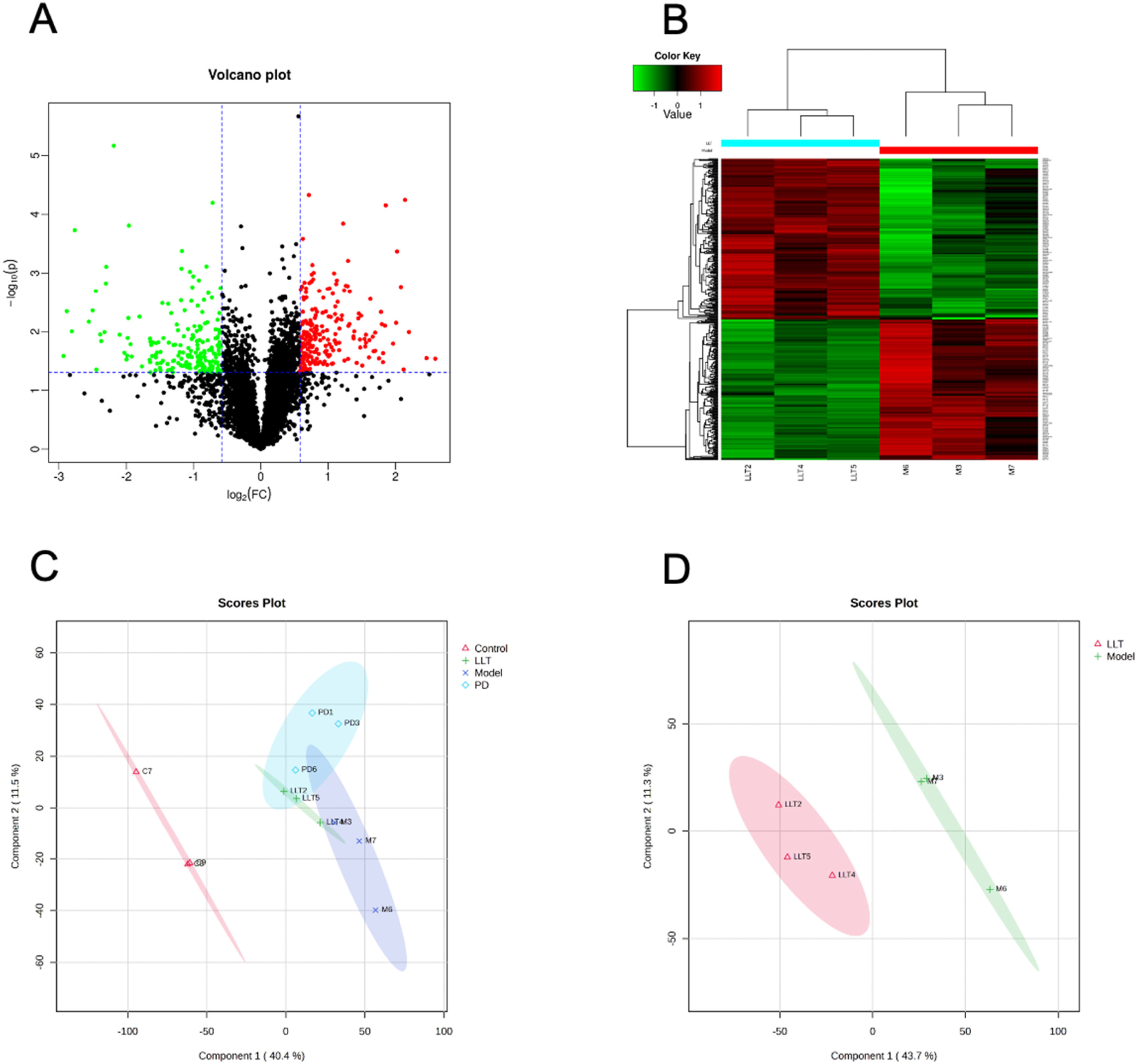
Zhang L. et al. Heliyon. 2024.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Label-Based Protein Quantitative Service—iTRAQ, TMT, SILAC
How to order?







