Histone PTM Data Analysis Service
Histone post-translational modifications (PTMs) are essential molecular switches that dynamically regulate chromatin architecture and gene expression. These covalent chemical modifications—including acetylation, methylation, phosphorylation, ubiquitination, and sumoylation—occur on histone tails and influence DNA accessibility, transcriptional activity, and chromatin remodeling. As such, histone PTMs serve as key epigenetic markers in cellular differentiation, development, immune regulation, and disease pathogenesis. The combinatorial nature of histone modifications, often referred to as the "histone code," represents a complex regulatory language. However, deciphering this code requires highly sensitive detection, site-specific mapping, and advanced computational interpretation. While experimental techniques such as mass spectrometry (MS) offer quantitative and site-level insights, extracting meaningful biological conclusions from raw data remains a major bottleneck for many researchers.
Service at MtoZ Biolabs
MtoZ Biolabs offers a dedicated Histone PTM Data Analysis Service which combines deep expertise in proteomics, epigenetics, and computational biology. This service supports MS-based histone PTM datasets, providing robust pipelines for data preprocessing, PTM identification, statistical analysis, and functional annotation. At MtoZ Biolabs, we bring extensive experience in large-scale proteomics and advanced histone PTM data analysis. We tailor our services to meet the unique needs of each project, offering customized data analysis and visualizations such as data tables, stacked bar charts, interactive volcano plots, heatmaps, and more. Whether your goal is to identify novel epigenetic markers or to compare histone PTMs across conditions, MtoZ Biolabs delivers a powerful solution tailored to your research objectives.
If you are not satisfied with the quality or results of your existing data, we also offer an all-in-one Histone Post-Translational Modification (PTM) Proteomics Service—from sample preparation and mass spectrometry analysis to comprehensive data interpretation—to ensure accurate identification of modification sites and high-quality data output.
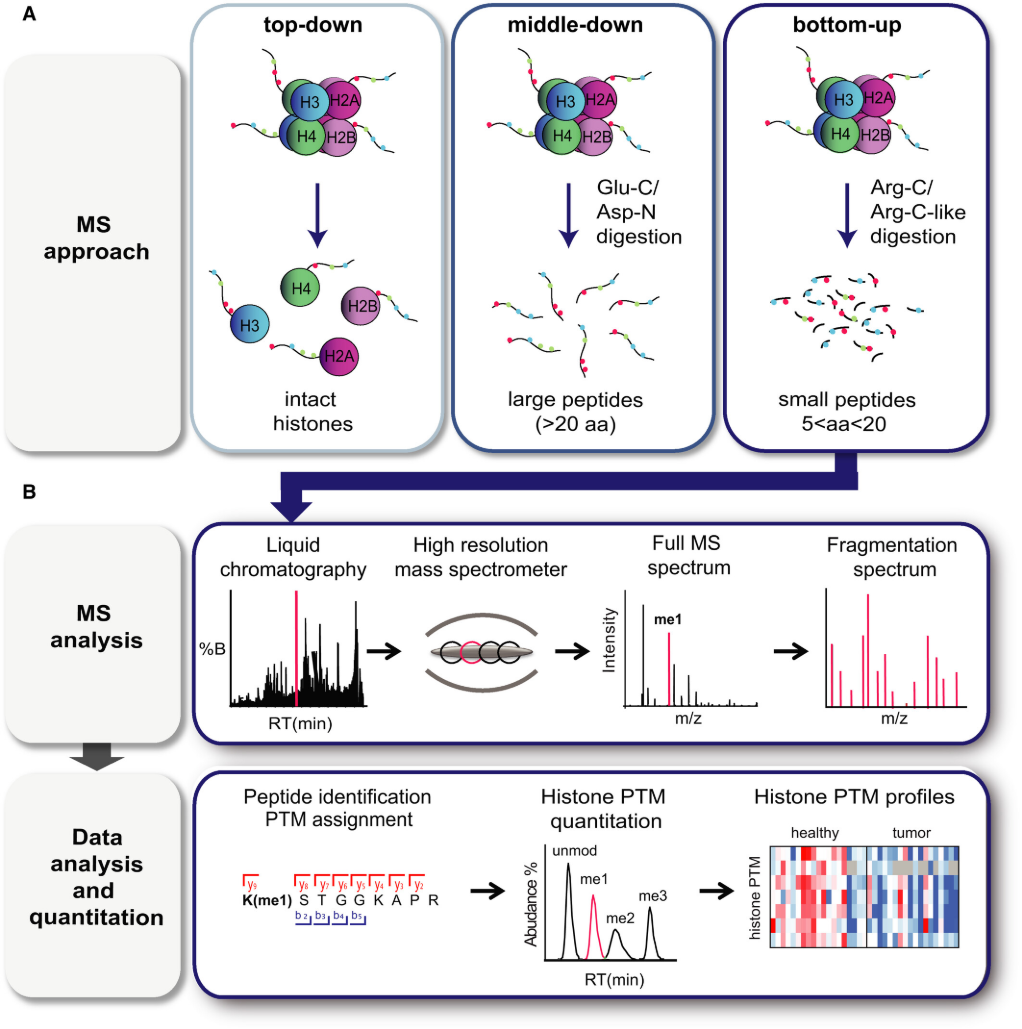
Figrue 1. MS‐Based Analysis of Histone PTMs. (A) Scheme Summarizing the Three MS‐Based Approaches that can be Used for Histone PTM Analysis. (B) Shows a General Bottom‐Up MS Workflow
Service Advantages
☑️High-Quality Data Interpretation: Accurate site-level identification and biologically meaningful annotation.
☑️Customizable Analysis Pipelines: Tailored workflows for different datasets, research goals, and experimental designs.
☑️Fast Turnaround: The workflow from data preprocessing to report generation is efficient, saving your time and effort.
☑️Experienced Bioinformatics Team: Proven track record in large-scale proteomics and epigenetics data interpretation.
Applications
1. Epigenetic Regulation Studies
Profile histone modification landscapes to understand chromatin state changes and gene expression regulation during development, differentiation, or reprogramming.
2. Developmental Biology
During embryogenesis and tissue development, histone modification patterns dynamically shape gene expression programs. By quantifying PTM changes at key loci, our service supports studies on lineage specification, morphogen gradients, and epigenetic memory.
3. Disease Epigenomics
Aberrant histone PTMs are frequently associated with oncogenesis, inflammation, and metabolic disorders. We provide comparative PTM analysis across disease vs. control conditions to identify diagnostic or therapeutic targets.
4. Drug Mechanism Research
Histone-modifying enzymes are common drug targets. Our service enables precise evaluation of compound-induced changes in histone marks, aiding in drug screening, target validation, and mechanistic studies.
Deliverables
1. Raw Data
Original mass spectrometry files or sequencing reads, as received and quality-checked.
2. PTM Site Identification
A list of confidently identified histone PTM sites with residue-level localization and modification types (e.g., acetylation, methylation states).
3. Quantitative Analysis
Normalized intensity data, fold change calculations, and statistical significance results across sample groups.
4. Functional Annotation & Pathway Analysis
Gene ontology enrichment, pathway mapping (e.g., KEGG, Reactome), and histone mark function annotation based on current databases and literature.
5. Comprehensive Analysis Report
A complete technical summary covering data processing methods, quality metrics, biological insights, and visual representations (e.g., volcano plots, heatmaps, bar charts).
With advanced analytical pipelines and expert interpretation, MtoZ Biolabs is committed to helping you transform complex histone PTM data into meaningful biological discoveries. Contact us today to discuss your project or request a custom proposal.
Related Services
Ubiquitylomics Data Analysis Service
How to order?







