ADP-Ribosylation Proteomics Service
ADP-Ribosylation Proteomics Service is a research service based on a high-resolution mass spectrometry platform, combining a variety of enrichment technologies and stable modification analysis strategies to perform global identification, site localization and relative quantification of protein ADP-ribose modification.
ADP-ribosylation is a type of post-translational modification catalyzed by the ADP-ribosyltransferase family (ARTs, also known as PARPs), in which an ADP-ribose group from NAD⁺ is covalently attached to specific sites on proteins (such as Arg, Glu, Asp, Ser, Cys, Lys, Tyr residues). This modification can exist as either mono-ADP-ribosylation (MARylation) or poly-ADP-ribose chain modification (PARylation), both of which are highly dynamic and reversible.
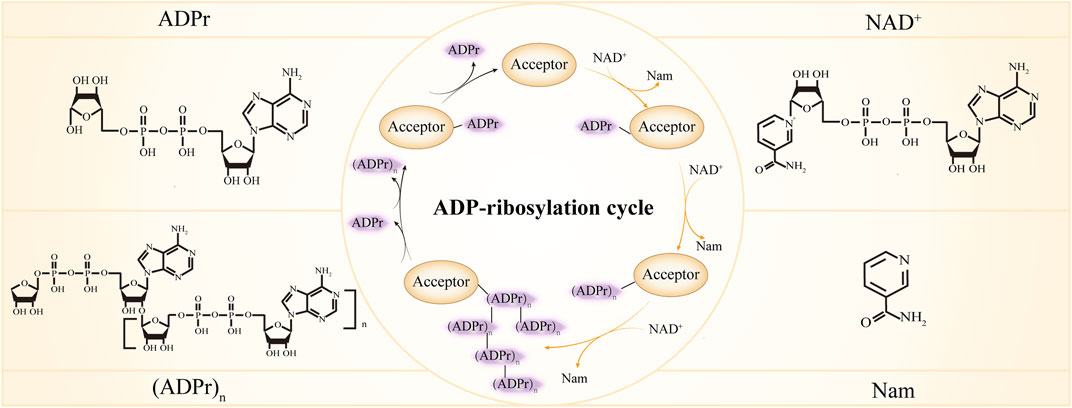
Li P. et al. Front Cell Dev Biol. 2022.
ADP-ribosylation is widely involved in processes such as DNA damage repair, chromatin remodeling, RNA metabolism, stress response, and viral infection regulation. Abnormal changes in ADP-ribosylation are closely associated with cancer, neurodegenerative diseases, autoimmune disorders, and inflammatory lesions. With the successful application of PARP inhibitors and other targeted therapies, the study of this modification has increasingly attracted attention in both basic and translational medicine. However, due to its complex chemical nature, low abundance, and structural instability, the systematic identification and quantification of ADP-ribosylation have long faced technical challenges. In recent years, with the development of high-resolution mass spectrometry and specific enrichment methods, ADP-ribosylation proteomics has become a key strategy for the comprehensive analysis of this modification.
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs offers the ADP-Ribosylation Proteomics Service to highly sensitively identify and locate ADP-ribosylation modifications on proteins, distinguish between monomeric and polymeric modification types, and quantitatively evaluate their dynamic changes under different conditions, helping researchers to systematically analyze the regulatory mechanism of this modification in processes such as DNA repair, stress response, viral infection, and tumorigenesis.
Analysis Workflow
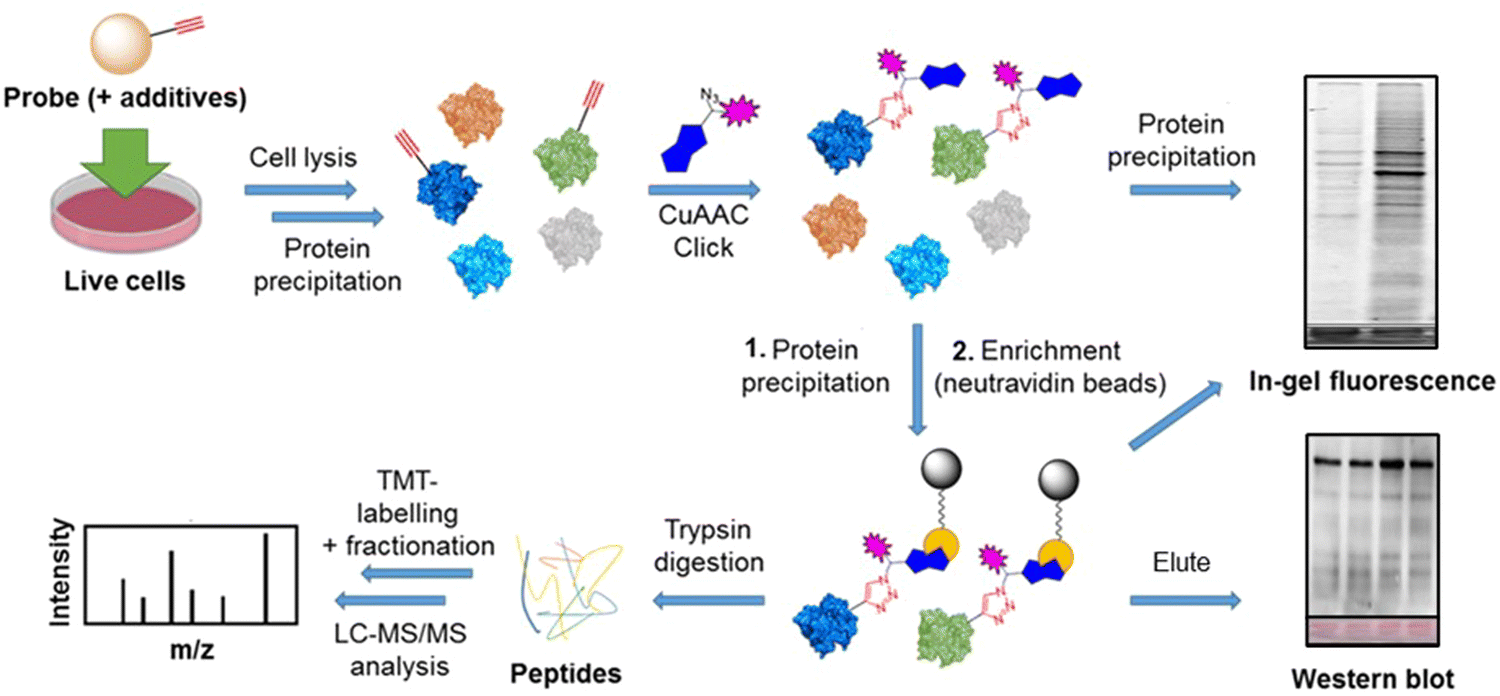
Draganov SD. et al. RSC Chem Biol. 2024.
The analytical workflow of ADP-Ribosylation Proteomics Service includes the following key steps:
1. Sample Preparation
Evaluate the sample, followed by cell lysis and protein extraction.
2. Enzymatic Digestion
Use trypsin or a multi-enzyme strategy (e.g., Lys-C) for digestion to generate peptides, while optimizing reaction conditions to retain modifications.
3. Modification Enrichment
Select enrichment strategies based on project needs, such as Af1521 affinity purification, antibody enrichment, or chemical modification capture, to isolate modified peptides.
4. LC-MS/MS Detection
Utilize a high-resolution Liquid Chromatography-Mass Spectrometry (LC-MS/MS) system to acquire full-scan and fragmentation data of peptides.
5. Data Analysis
Use specialized software for peptide alignment, modification site localization, and quantitative comparison, along with pathway enrichment and functional annotation via database integration.
6. Result Delivery
Provide a comprehensive report, including protein and modification site tables, quantitative variation spectra, enrichment analysis graphs, and MS/MS spectra.
Service Advantages
1. Precise Modification Site Identification: Utilizing advanced mass spectrometry platforms, we support high-confidence identification of modification sites and sensitive detection of low-abundance modifications.
2. Support for Dynamic Quantitative Analysis: Through label-free quantification or TMT/iTRAQ labeling, we enable dynamic comparison of modification levels under different treatment conditions, aiding mechanism analysis and efficacy evaluation.
3. Multi-Strategy Enrichment: Combining Af1521 affinity purification, antibody enrichment, and chemical derivatization, we accurately capture ADP-ribosylation modifications, providing comprehensive coverage of both MARylation and PARylation modifications.
4. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
5. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all ADP-Ribosylation Proteomics data, providing clients with a comprehensive data report.
Sample Submission Suggestions
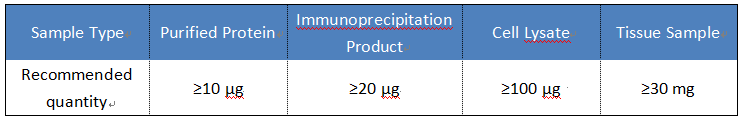
It is recommended that samples be submitted in a cryogenically frozen state (–80°C); liquid samples need to be sealed and aliquoted and transported on dry ice to avoid repeated freezing and thawing.
You are welcome to communicate your specific needs with the MtoZ Biolabs technical team before submitting your samples. We can provide customized sample processing suggestions and pre-treatment services.
Applications
Application examples of the ADP-Ribosylation Proteomics Service:
DNA Damage and Repair Mechanism Research: Gain deeper insights into the PARP regulatory network and chromatin modification patterns.
Cancer Signaling Pathway Research: Analyze the mechanisms of ADP-ribosylation dysregulation in tumor cells and its correlation with drug resistance.
Antiviral and Immune Regulation Research: Explore the role of ADP-ribosylation in host antiviral responses and immune evasion.
PARP Targeted Drug Mechanism Validation: Assess the dynamic changes in the ADP-ribosylome before and after treatment with PARP inhibitors.
Cellular Stress and Metabolic Response Research: Analyze the global changes in ADP-ribosylation under conditions of oxidative stress, heat shock, and other stress responses.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Related Services
Post-Translational Modifications Proteomics Service
Deamidation Proteomics Service
How to order?







