Polyglutamylation Proteomics Service
Polyglutamylation Proteomics Service refers to a service that utilizes high-resolution mass spectrometry combined with modification-specific enrichment techniques to systematically identify, localize, and quantitatively analyze polyglutamylation modification sites, chain length isoforms, and their dynamic changes in proteins. This service enables precise localization and spectral interpretation of polyglutamylation chain lengths, polymodification sites, and structural isomers, facilitating dynamic analysis of complex modification states.
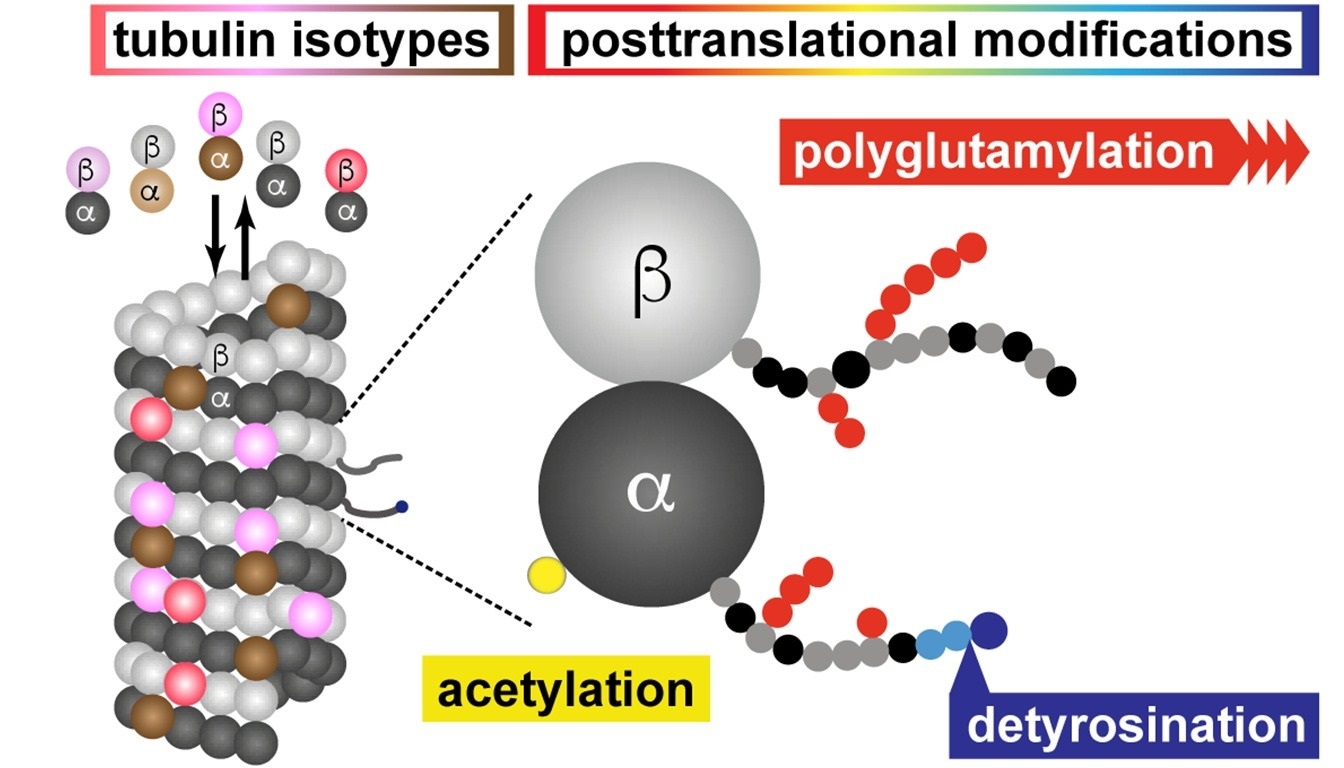
Bodakuntla S. et al. Neuroscience letters. 2021.
Polyglutamylation is a specific type of post-translational modification (PTM), in which multiple glutamate residues are sequentially attached via amide bonds to the γ-carboxyl group of a glutamine or glutamate side chain on a target protein. This modification is highly variable and includes both the initiation glutamylation site on the protein backbone (initiation Glu) and variations in the length of the glutamate side chain extension. Polyglutamylation was first discovered on tubulin and is known to be broadly involved in regulating cytoskeletal dynamics, protein–protein interactions, signaling pathways, ciliary function, and the pathogenesis of neurodegenerative diseases. It has become a key focus in studies of cell assembly dynamics, neurological dysfunction, and cancer cell migration mechanisms.
Relying on high-resolution mass spectrometry and other platforms, MtoZ Biolabs provides Polyglutamylation Proteomics Service that can accurately identify and locate polyglutamylation modification sites in proteins, analyze the length changes and modified isomers of polyglutamylation chains, support quantitative comparisons under multiple conditions, and help researchers deeply explore the key role of this modification in microtubule stability, neural function, signal regulation, and disease mechanisms.
Analysis Workflow
Brief Workflow of Polyglutamylation Proteomics Service:
1. Sample Preparation
Total proteins are extracted and quantified to ensure that sample quality meets the requirements for downstream analysis.
2. Proteolytic Digestion
Proteins are digested into peptides using proteases, with polyglutamylated regions retained.
3. Enrichment of Modified Peptides
Polyglutamylated peptides are enriched using anti-polyglutamylation antibodies or immunoprecipitation strategies.
4. Mass Spectrometry Detection
Enriched peptides are analyzed using high-resolution LC-MS/MS to acquire MS1 and MS2 spectra.
5. Data Analysis and Annotation
polyglutamylation modification sites and chain lengths are localized using modification identification algorithms, followed by functional annotation and visualized data output.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Polyglutamylation Proteomics Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Polyglutamylation Proteomics Service data, providing clients with a comprehensive data report.
Applications
Application Examples of Polyglutamylation Proteomics Service:
Mechanistic Studies of Neuronal Microtubule Regulation
Identify polyglutamylation patterns on tubulin and their association with axonal transport defects to explore novel mechanisms in neurodegenerative diseases.
Chromatin Stability and DNA Repair Research
Analyze how polyglutamylation modifications on histones or repair factors affect chromosome integrity and DNA repair pathways.
Analysis of Cancer Cell Migration and Adhesion Regulation
Compare polyglutamylation modification changes under different tumor conditions to uncover regulatory markers of cell motility and invasiveness.
TTLL and CCP Functional Studies and Substrate Profiling
Systematically investigate the substrate spectra of polyglutamylation-related enzyme families and their chain length–specific regulatory mechanisms.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
FAQ
Q. Which Proteins Can be Analyzed for Polyglutamylation Using this Service?
This service is suitable for detecting polyglutamylation on proteins with modification potential, especially tubulin, histone variants, enzyme substrates (such as those from the TTLL/CCP families), and proteins involved in signaling or cytoskeletal regulation. We also support discovery-based studies for unknown polyglutamylated substrates.
Q. Can Different Polyglutamylation Chain Lengths be Distinguished? How Many Chain Length Isoforms Can be Identified?
Yes. Using high-resolution mass spectrometry and multi-stage fragmentation data, we can identify and distinguish polyglutamylation chain length isoforms containing at least 1–8 glutamate units. The specific resolution capacity depends on sample complexity and modification abundance.
Q. Does the Service Support Combined Analysis of Polyglutamylation with Other PTMs?
Yes. Upon request, we can simultaneously identify polyglutamylation along with other PTMs such as phosphorylation, acetylation, deacylation, and ubiquitination. We can also assess their co-occurrence on the same peptide or protein to reveal potential cross-talk mechanisms among PTMs.
Case Study
This study systematically analyzed the regulatory network of polyglutamylation on tubulin. By integrating anti-polyglutamylation antibody enrichment, mass spectrometry analysis, and functional proteomics, the researchers identified and characterized a novel regulatory complex, C11ORF49. This factor forms a complex (TPGC) with TTLL glutamylases and CCP deglutamylases to regulate the level and spatial distribution of polyglutamylation on microtubules. The results demonstrated that complex deficiency leads to severe dysregulation of tubulin polyglutamylation, thereby compromising nuclear morphology stability, delaying cilia disassembly, and affecting cytoskeletal architecture by modulating the dynamic balance between microtubules and actin filaments.
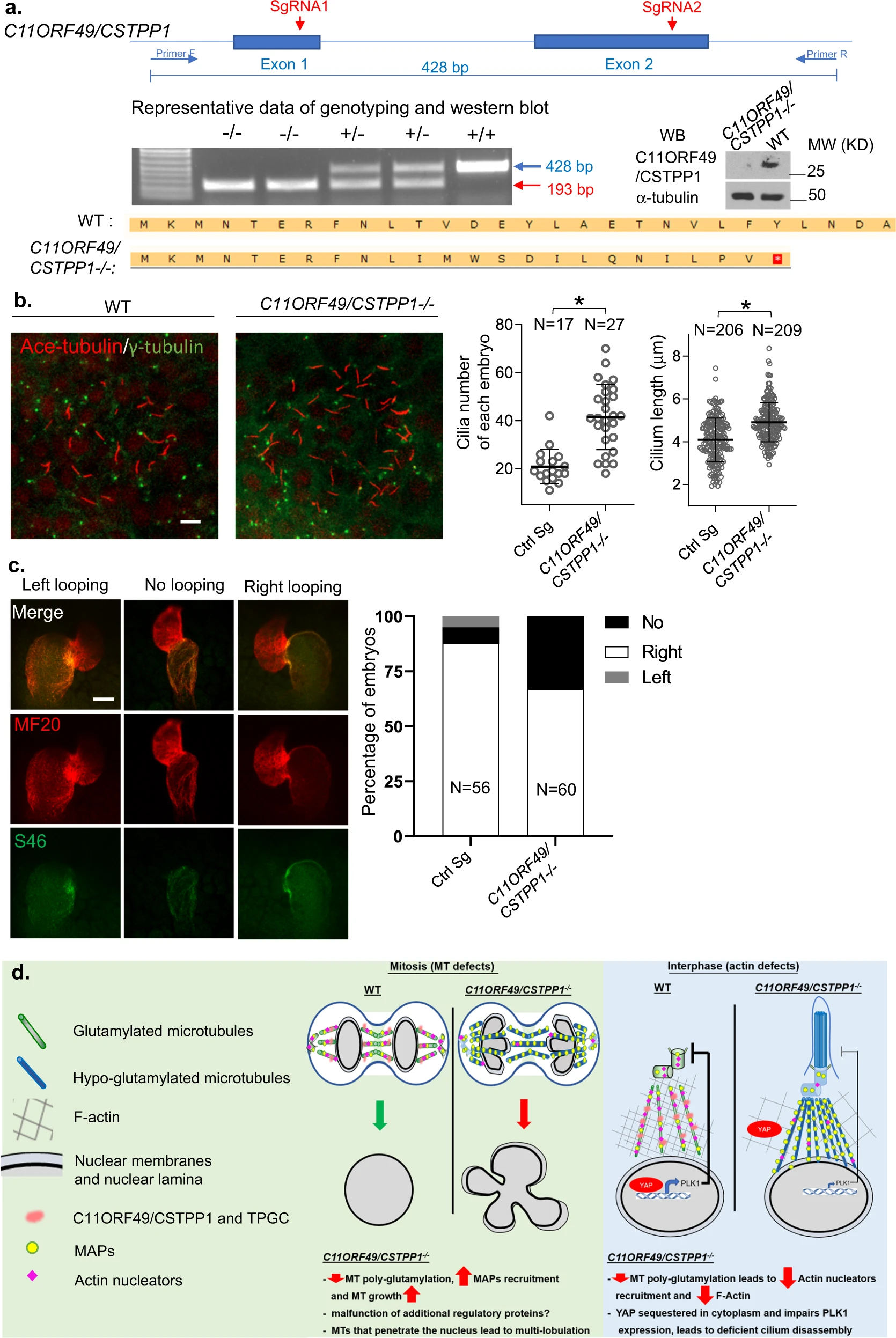
Wang L. et al. Cell Res. 2022.
How to order?







