TMT Mass Spectrometry Service
TMT-based mass spectrometry is a high-throughput, high-precision proteomics analysis technique that uses specially designed isotopic chemical reagents (TMT tags) to label proteins or peptides. It quantifies the relative abundance of proteins across different samples by measuring the intensity of reporter ions released during fragmentation in the mass spectrometer. In TMT mass spectrometry, a set of TMT reagents includes tags with identical overall mass. The chemical structure of each tag consists of a reporter group, a mass-balancing group, and a peptide-reactive group. The peptide-reactive group covalently binds to the free amino groups on the N-terminus or lysine side chains of peptides, enabling the labeling of all peptides with TMT tags. Different TMT tags are distinguished by unique reporter groups, while the mass-balancing group ensures all TMT tags have identical overall molecular weight. During tandem mass spectrometry (MS/MS), the reporter groups are released through fragmentation and generate reporter ion peaks in the low-mass region of the spectrum. The intensity of these reporter ions reflects the relative abundance of a specific peptide across different samples. TMT mass spectrometry service allows the labeling of multiple samples, which are then analyzed simultaneously in a single run, distinguishing the protein expression levels in each sample based on the isotopic labels.
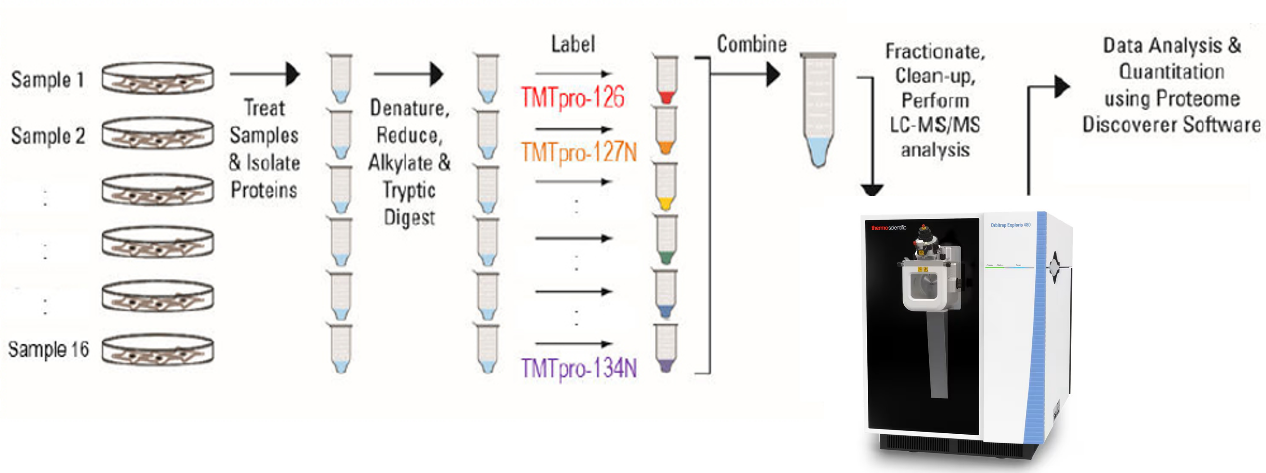
Figure 1. Basic Principle of TMT Mass Spectrometry.
Service Advantages
MtoZ Biolabs provides proteomics services based on TMT mass spectrometry technology. Compared to conventional label-free methods, TMT mass spectrometry offers multiple advantages:
1. High-Throughput Analysis
TMT mass spectrometry service allows simultaneous labeling and analysis of multiple samples. A single TMT reagent kit can process up to 16 or more samples, accelerating workflows, improving data collection efficiency, and minimizing inter-experimental variability.
2. High-Precision Quantification
TMT labels utilize isotopic tags, enabling precise differentiation of peptides from various samples during mass spectrometry analysis. The intensity of the reporter ions accurately reflects the relative abundance of proteins, ensuring precise quantification.
3. Wide Dynamic Range
TMT mass spectrometry service is suitable for detecting proteins with large abundance differences. Even low-abundance proteins can be effectively detected and quantified after TMT labeling, covering a broader range of protein expression.
4. High Sensitivity
TMT labeling enhances ion yield, making TMT mass spectrometry highly sensitive for detecting low-abundance proteins. This allows for identifying rare or low-expression proteins that are difficult to detect with traditional methods.
5. Reliable Data
By integrating isotopic labeling and mass spectrometry, TMT mass spectrometry service provides stable and reliable data with excellent reproducibility, reducing experimental errors and false positives.
6. Broad Applications
TMT mass spectrometry has extensive applications in biomedical research, including disease studies, drug development, and biological investigations. It aids in uncovering protein expression differences, identifying potential biomarkers, and discovering drug targets.
Sample Submission Suggestions
1. Sample Types
We accommodate a variety of sample types, including gel bands, protein solutions, solid powders, cells, animal and plant tissues, and metabolites.
2. Sample Amount
The required quantity varies depending on the sample type and project. Please consult us for specifics.
Note: For special requirements or sample preparation assistance, please contact us.
Applications
1. Disease Research
TMT mass spectrometry service facilitates studying protein expression changes in different biological states, providing insights into disease mechanisms. By comparing diseased and healthy samples, researchers can identify protein expression changes associated with specific diseases, aiding in biomarker discovery.
2. Drug Development
During drug development, TMT mass spectrometry can identify drug targets and evaluate drug efficacy. By comparing samples before and after treatment, the molecular targets and mechanisms of action can be revealed, supporting pharmacodynamics studies and novel drug discovery.
3. Cell Signaling Research
TMT mass spectrometry service can investigate protein expression changes in cells under specific stimuli (e.g., hormones, drugs, stress conditions), uncovering the regulatory mechanisms of signaling pathways.
4. Differential Protein Expression Analysis
Using TMT mass spectrometry to analyze protein expression differences across physiological states, disease states, or drug treatments helps identify proteins linked to specific biological processes or conditions.
5. Post-Translational Modifications (PTMs)
Post-translational modification is a process in which proteins undergo chemical modifications after translation, such as phosphorylation, acetylation, methylation, etc. TMT mass spectrometry can identify and quantify the types and levels of post-translational modifications in different samples, helping to deeply understand the regulatory mechanism of protein function and its association with diseases.
6. Protein Interaction Network Construction
Analyzing protein complexes or interaction networks through TMT mass spectrometry service can reveal signaling pathways, metabolic pathways and other biological processes between proteins, providing important clues for studying the function and regulation of biological systems.
Case Study
1. Using TMT mass spectrometry to Investigate Cervical Cancer Mechanisms
Cervical cancer is the second most common gynecological malignancy, and its pathogenesis remains unclear. TMT mass spectrometry combined with bioinformatics tools was used to analyze exfoliated cervical cells from normal and cervical cancer groups to establish a cancer-specific proteome and identify key proteins associated with cervical cancer development. A total of 351 differentially expressed proteins were identified in the cervical cancer group compared with the normal group, including 247 upregulated and 104 downregulated proteins. Gene ontology functional annotation showed that differentially expressed proteins were mainly involved in unicellular-multicellular biological processes, multicellular biological processes, and negative regulation of biological processes. These proteins play a role in the molecular functions of extracellular membrane-binding organs of cellular components, cellular exosomes, protein binding, structural molecular activity, and enzyme binding. These differentially expressed proteins are mainly involved in signaling pathways such as PI3K-Akt, extracellular matrix receptor interaction, complement and coagulation cascades. In particular, peroxisome proliferator-activated receptor 2 may be involved in cervical tumorigenesis by inhibiting apoptotic signaling. This study determined that proteins in the cervical cancer group were altered both qualitatively and quantitatively, and a total of 351 differentially expressed proteins were identified. The functions and signaling pathways of these differentially expressed proteins lay a theoretical foundation for elucidating the molecular mechanisms of cervical cancer.
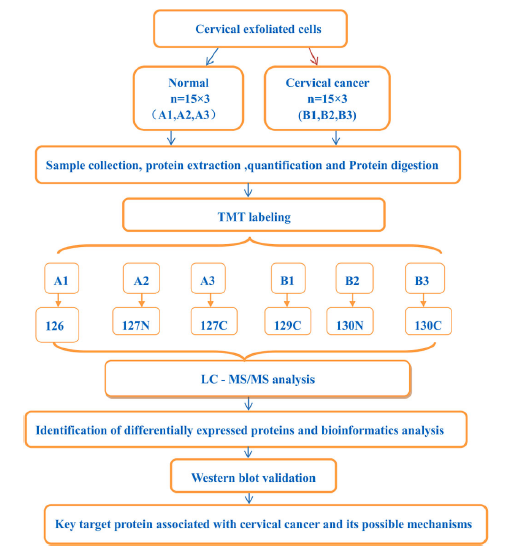
Xu, D Q. et al. Journal of Proteomics, 2022.
Figure 2. Flow Chart for Studying the Pathogenesis of Cervical Cancer Using TMT Mass Spectrometry.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
TMT/iTRAQ Labeling-Based Quantitative Service
SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture)-Based Quantitative Service
How to order?







