Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service
Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) is a quantitative proteomics technique used to measure protein abundance, dynamics, and post-translational modifications (PTMs) in living cells. Stable Isotope Labeling with Amino Acids in Cell Culture method enables precise and reproducible quantification of differential protein abundance, making it indispensable for studying protein dynamics in complex biological systems.
The Stable Isotope Labeling with Amino Acids in Cell Culture method involves culturing two groups of cells in distinct media: the "light" medium, which contains naturally occurring amino acids (such as 12C or 14N), and the "heavy" medium, which contains stable isotope-labeled amino acids (such as 13C or 15N). Over 5-6 cell divisions, the labeled amino acids are incorporated into newly synthesized proteins, resulting in proteins with a mass shift compared to the unlabeled proteins in the light medium. Once labeling is complete, equal amounts of labeled and unlabeled cells or protein extracts are mixed. These mixtures are then digested into peptides using trypsin. The resulting peptides are analyzed via mass spectrometry (MS), which differentiates labeled peptides from unlabeled ones based on their mass shift. The quantitative analysis of Stable Isotope Labeling with Amino Acids in Cell Culture relies on comparing the signal intensities of labeled and unlabeled peptides, allowing for the accurate determination of relative protein abundance in the sample.
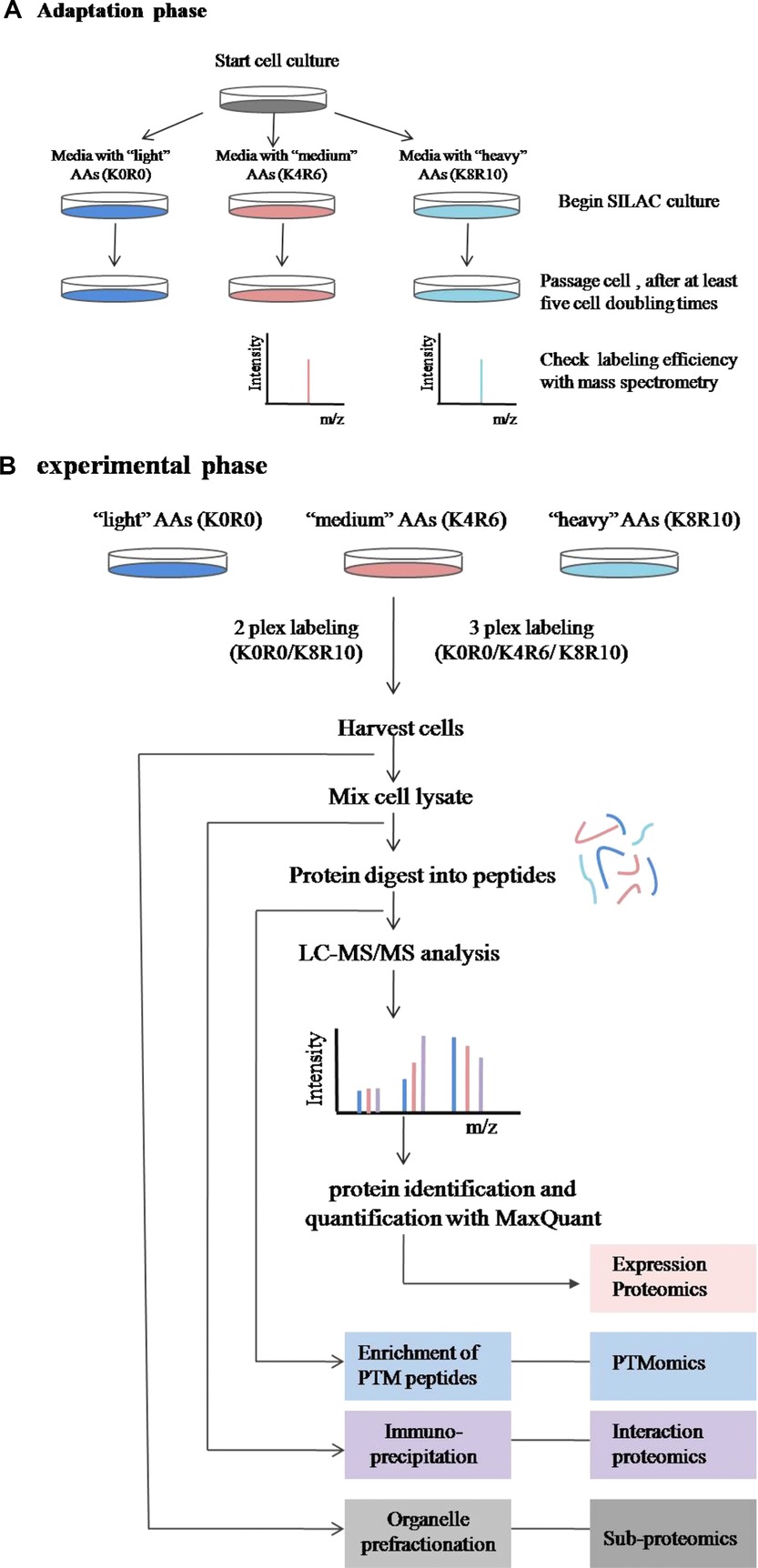
Chen, X. et al. Proteomics. 2015.
Figure 1. Workflow for Quantitative Proteomic Experiments Using Stable Isotope Labeling with Amino Acids in Cell Culture
Service at MtoZ Biolabs
MtoZ Biolabs offers a comprehensive Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service that enables researchers to quantify protein expression under different experimental conditions. The Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service requires extended cell culture time to ensure complete incorporation of labeled amino acids into the proteome. This careful labeling process ensures that the resulting data is accurate and reproducible, providing deep insights into protein dynamics and cellular mechanisms. Free project evaluation, welcome to learn more details!
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. High Labeling Efficiency: Our Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service achieves labeling efficiencies of over 99%, ensuring accurate and reliable protein quantification with minimal variation.
3. Minimal Sample Requirements: Only tens of micrograms of protein are needed per sample, making this service ideal for studies with limited sample amounts.
4. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service data, providing clients with a comprehensive data report.
5. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
Applications
The Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service offers several powerful applications for proteomics research:
1. Quantitative Protein Expression
Monitor protein expression changes under various conditions, providing insights into disease mechanisms, drug responses, or environmental effects.
2. Protein Turnover
Track protein synthesis and degradation rates by incorporating stable isotope-labeled amino acids, allowing for detailed analysis of protein turnover.
3. Post-Translational Modifications (PTMs)
Investigate dynamic changes in PTMs, such as phosphorylation or acetylation, in response to different cellular stimuli.
4. Protein-Protein Interactions (PPIs)
Identify and quantify protein-protein interactions (PPIs) by distinguishing true interacting partners from non-specific contaminants.
Case Study
Using SILAC to Investigate MYH9-Actin Interaction in Cancer Therapy
In this study, the Stable Isotope Labeling with Amino Acids in Cell Culture method was used to investigate the role of the MYH9-actin molecular motor in mitochondrial fission and its potential as an anti-cancer target. By coupling SILAC with pulldown analysis, HSPA9 was identified as a crucial adaptor protein linking MYH9-actin to mitochondrial fission. Treatment with the small-molecule J13 disrupted the MYH9-actin interaction, causing mitochondrial imbalance and inhibiting cancer cell proliferation and migration. This study demonstrates the utility of Stable Isotope Labeling with Amino Acids in Cell Culture Analysis Service in identifying key molecular interactions for therapeutic targeting in cancer research.
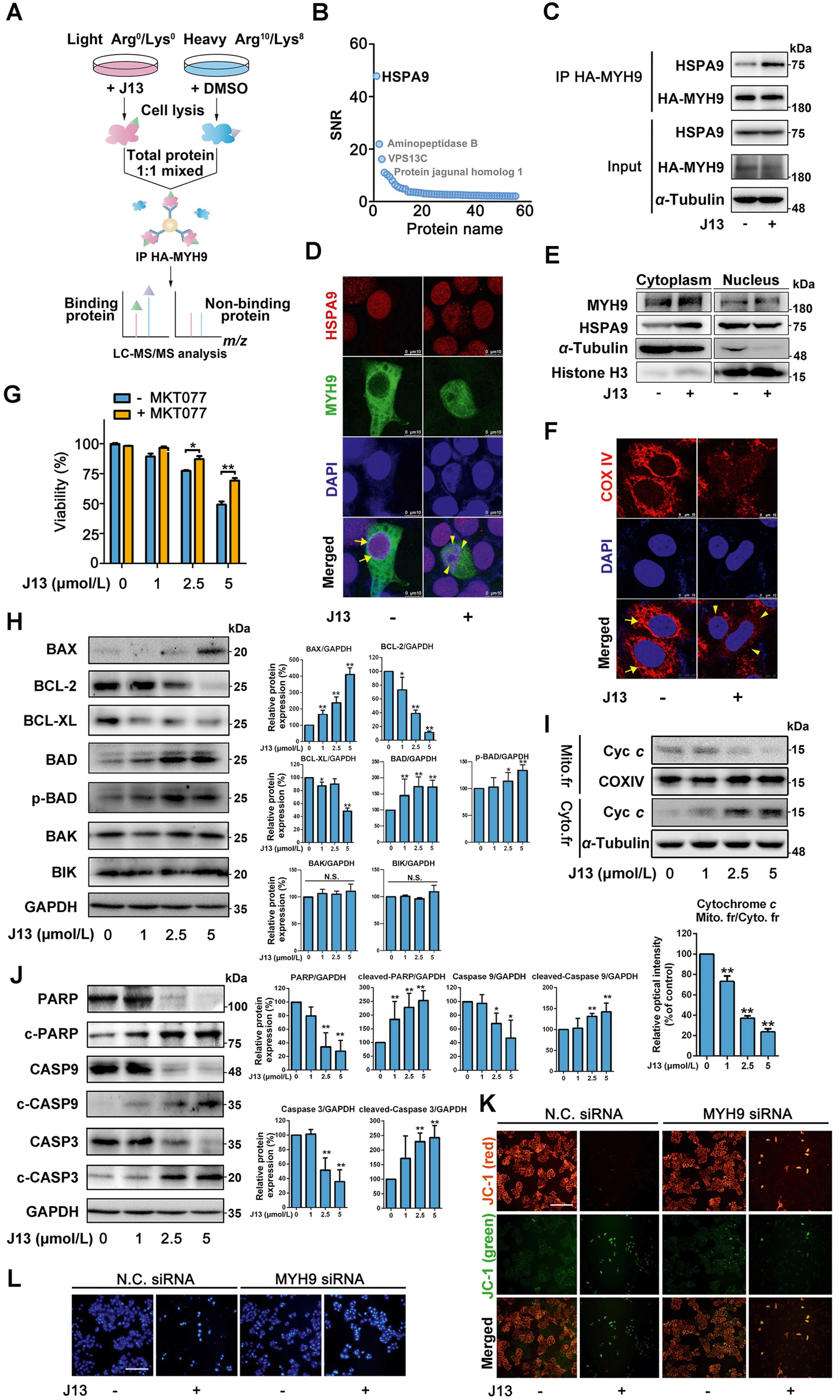
Qian, Y. et al. Acta Pharm Sin B. 2021.
FAQ
Q: What factors influence the labeling efficiency in Stable Isotope Labeling with Amino Acids in Cell Culture, and how can they be optimized?
Labeling efficiency in Stable Isotope Labeling with Amino Acids in Cell Culture is influenced by factors such as cell growth rate, the medium composition, and the duration of labeling. Optimizing these factors involves adjusting cell culture conditions, ensuring adequate isotope availability, and allowing sufficient time for complete incorporation of labeled amino acids.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
SILAC/Dimethyl Quantitative Proteomics Service
iTRAQ/TMT/MultiNotch Quantitative Proteomics Service
SILAC Based Co-IP-MS for Protein Interaction Analysis Service
How to order?







