SILAC-Based Peptidomics Analysis Service
SILAC (stable isotope labeling by amino acids in cell culture) is a metabolically based quantitative proteomics technique widely applied in peptidomics research. Its fundamental principle involves introducing light and heavy isotope-labeled amino acids into cell culture, enabling relative quantification of peptides across different experimental groups. This method requires no additional chemical labeling, offers high labeling efficiency, and minimizes inter-sample variability, making it especially suitable for precise studies of small peptides involved in dynamic processes and signaling pathways.
The SILAC-based peptidomics analysis service is widely used in areas such as cell communication mechanism research, signal transduction regulation, functional peptide screening, and differential analysis of disease-related peptides. It demonstrates distinct advantages particularly in cancer biomarker discovery, drug mechanism investigation, and neuropeptide research.
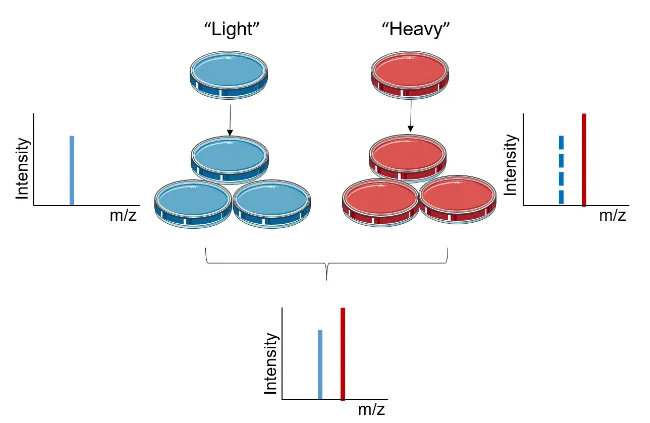
Chen, X. et al. Proteomics, 2015.
Figure 1. Experimental Principle of SILAC.
Services at MtoZ Biolabs
Based on a high-resolution orbitrap mass spectrometry system, MtoZ Biolabs offers the SILAC-based peptidomics analysis service focusing on quantitative analysis of peptides. This service employs stable isotope-labeled amino acids to enable precise comparison of peptide expression differences between treatment groups. It is suitable for analyzing dynamic changes in functional peptides, signal peptides, and neuropeptides, delivering highly reproducible and high-resolution quantitative spectral data to support the elucidation of peptide biological functions in cellular responses and regulatory processes.
Analysis Workflow
1. SILAC Labeling Cell Culture
Cells are cultured in media containing light or heavy isotope-labeled amino acids to achieve stable isotope labeling of endogenous peptides.
2. Cell Lysis and Endogenous Peptide Extraction
Cells are lysed without the addition of proteases, and natural endogenous peptides are enriched.
3. Mixing of Isotope-Labeled Samples
Equal amounts of peptide extracts from light- and heavy-labeled groups are mixed and processed together to eliminate batch-to-batch variation and improve quantification accuracy.
4. Liquid Chromatography–Mass Spectrometry (LC-MS/MS)
Using a high-resolution Orbitrap mass spectrometry platform, peptides in the mixed samples are separated and detected, enabling highly sensitive data acquisition.
5. Data Processing and Differential Analysis
Specialized software is used to identify labeled peptides, calculate quantitative ratios, and perform statistical analysis to generate peptide expression profiles and differential results.
Service Advantages
1. Advanced Analytical Platform
Powered by high-resolution orbitrap mass spectrometry combined with stable isotope labeling strategies, this service enables highly sensitive identification and quantification of complex peptide samples.
2. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
3. High-Quality Data
SILAC labeling significantly reduces inter-batch variation, delivering highly consistent and reproducible relative quantification data suitable for rigorous scientific studies.
4. Customized Solutions
MtoZ Biolabs offers fully customized workflows encompassing cell culture, sample preparation, mass spectrometry analysis, and data processing to accommodate diverse research needs.
Applications
1. Proteolytic Regulation Studies
SILAC-based peptidomics analysis service enables systematic analysis of endogenous protein cleavage patterns under specific physiological or experimental conditions, uncovering mechanisms of protein degradation and proteolytic regulation.
2. Assessment of Cellular Functional States
By quantitatively comparing endogenous peptide changes under different treatment conditions, the service reflects cellular functional alterations and signaling regulation status.
3. Monitoring of Development and Differentiation Processes
SILAC-based peptidomics analysis service can be used to track dynamic changes in peptidome composition during cell or tissue development and differentiation, revealing stage-specific regulatory characteristics.
4. Fundamental Proteomics Research
This service supports in-depth understanding of peptidome composition and regulatory mechanisms, providing valuable data for protein function analysis and systems biology research.
FAQ
Q1: Does the Service Support the Analysis of Post-Translationally Modified (PTM) Peptides?
A1: Yes, but enrichment or screening strategies need to be designed for specific modifications, such as phosphorylation or acetylation. MtoZ Biolabs offers corresponding customized services.
Q2: Does SILAC Labeling Affect Cell State or Expression Profiles?
A2: Typically, it does not cause significant effects. The structural differences between light and heavy labeled amino acids are minimal and do not interfere with protein expression or cellular functions.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
How to order?







