Qualitative Proteomics
- Proteome Mapping: Establishes baseline proteomic profiles for specific tissues, species, or experimental conditions.
- Disease-Associated Protein Discovery: Identifies proteins with condition-specific expression in diseased tissues or body fluids, supporting downstream biomarker development.
- Sample Quality Control and Authentication: Verifies the composition and purity of bioproducts such as recombinant proteins, antibodies, and vaccines.
- Fundamental Biological Research: Characterizes protein expression patterns during cell-type-specific processes such as differentiation, activation, and apoptosis.
Qualitative Proteomics is an analytical approach that uses high-resolution mass spectrometry to identify proteins present in a sample. The primary goal is to determine which proteins exist within the sample and, through database matching, infer their peptide sequences, originating genes, and functional properties. This method does not involve quantitative comparison of protein abundance but focuses instead on protein diversity, structural features, post-translational modifications, and their associations with other biomolecules.
As a fundamental component of basic research, qualitative proteomics is dedicated to identifying and characterizing all proteins in a biological sample. It provides critical information for understanding cellular functions, uncovering disease mechanisms, and constructing biological pathway maps. Compared to quantitative proteomics, qualitative analysis emphasizes protein presence and composition, making it particularly suitable for exploratory studies, proteome mapping of new species, identification of proteins in complex mixtures, and discovery of previously uncharacterized proteins. It serves as an essential foundation for multi-omics integration, post-translational modification profiling, and biomarker discovery.
Services at MtoZ Biolabs
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs provides high-quality Qualitative Proteomics services to accurately identify protein composition in a wide range of biological samples. Our services cover protein extraction, enzymatic hydrolysis, mass spectrometry detection, database retrieval, and functional annotation, and are applicable to various research needs such as unknown protein identification, species protein spectrum construction, biomarker screening, and biological product component analysis. Combined with customized bioinformatics analysis, these services enable researchers to systematically analyze protein expression within their samples, laying a solid foundation for downstream quantitative studies and mechanistic insights.
MtoZ Biolabs also offers the following related services:
Large-Scale Protein Identification Service
Utilizing high-throughput mass spectrometry platforms for comprehensive protein identification in complex samples. Supports species-wide proteome profiling, tissue-specific expression analysis, and disease-related protein screening.
Protein Interaction Analysis Service
Based on strategies such as co-immunoprecipitation, pull-down assays, or crosslinking mass spectrometry, this service identifies direct and indirect protein-protein interactions, facilitating the elucidation of signaling pathways and functional networks.
Integrates proteomics approaches to systematically analyze PROTAC induced target protein degradation and ubiquitination patterns, providing data support for efficacy evaluation and target validation.
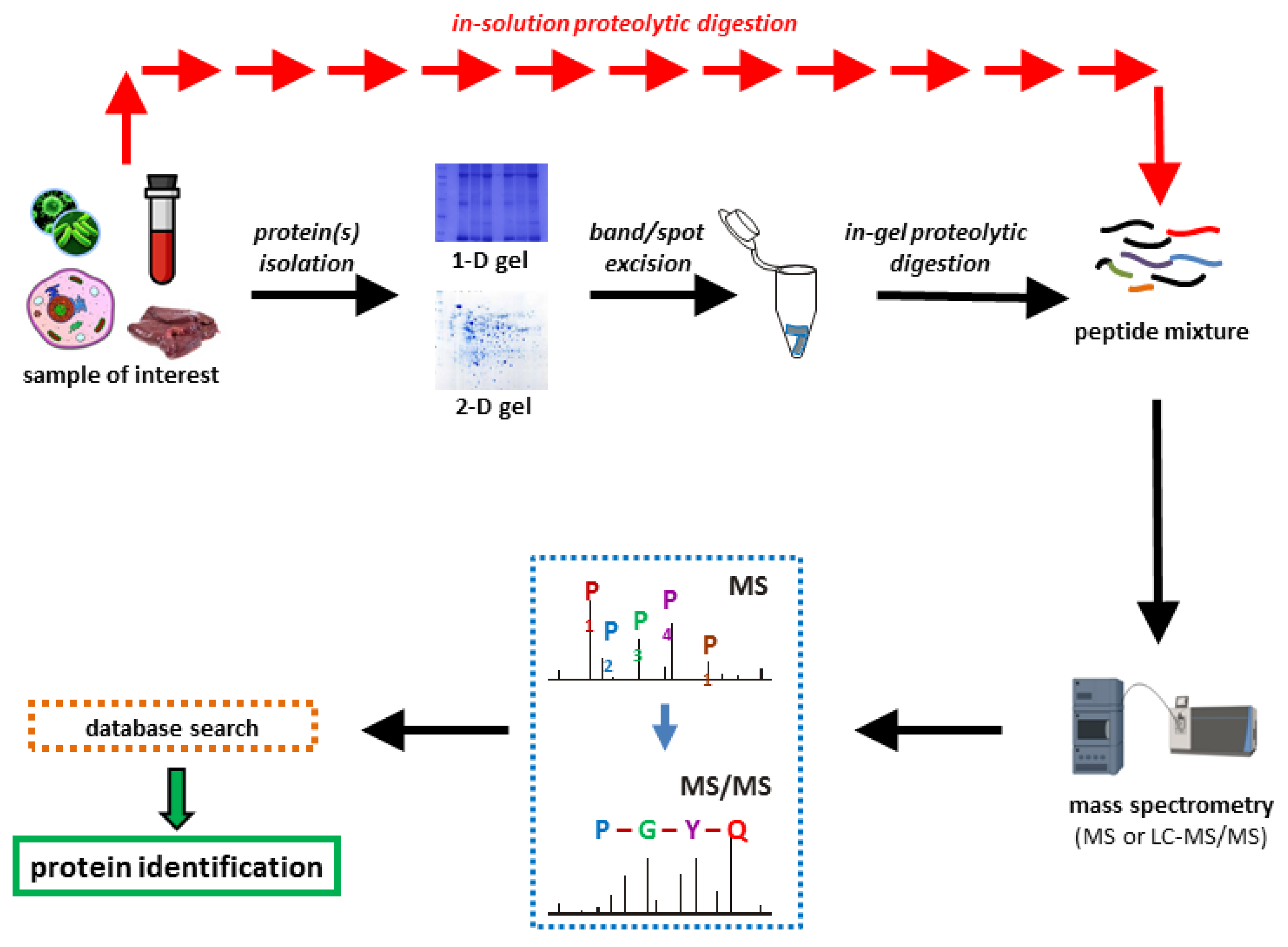
Dupree EJ. et al. Proteomes. 2020.
Figure 1. Workflow in a bottom-up proteomics experiment.
Analysis Workflow
The general workflow of Qualitative Proteomics is as follows:
1. Sample Preparation
Includes protein extraction, concentration measurement, enzymatic digestion, and necessary desalting and purification steps.
2. LC-MS/MS Analysis
Peptides are analyzed using a high-performance nano-liquid chromatography system coupled with a high-resolution mass spectrometry platform for data acquisition.
3. Database Searching
MS/MS data are searched against relevant protein databases to obtain protein and peptide identification results.
4. Result Filtering and Annotation
Quality control is performed to generate a high-confidence protein identification list, followed by functional annotation, classification, and statistical analysis.
5. Optional analysis module (customizable)
Includes post-translational modification (PTM) identification, subcellular localization prediction, protein interaction analysis, and exploratory analysis of differential expression trends.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Qualitative Proteomics platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Qualitative Proteomics data, providing clients with a comprehensive data report.
4. High Sensitivity and High-Throughput Identification: Capable of identifying thousands of proteins in complex samples with broad coverage and deep proteome profiling.
5. Compatible with Diverse Sample Types: Applicable to a wide range of biological materials including tissues, cells, body fluids, microorganisms, and biopharmaceutical products—flexibly supporting various research objectives.
Applications
Qualitative Proteomics is widely applied in life sciences and biomedical research, including but not limited to:
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
How to order?







