Large-Scale Protein Identification Service
Large-Scale Protein Identification Service refers to the systematic identification and analysis of a large number of proteins using high-throughput technology platforms. This service is designed to rapidly, accurately, and on a large scale identify and quantify various proteins from complex biological samples, primarily used indiscovery proteomics research. Large-scale protein identification mainly relies on modern mass spectrometry technologies, particularly liquid chromatography-tandem mass spectrometry (LC-MS/MS) systems, which can obtain identification information for a large number of proteins in a short period of time. LC-MS/MS separates protein samples via liquid chromatography, and then the mass spectrometer detects peptide ion fragments with high resolution, combining database searches for protein identification and quantification. The Large-Scale Protein Identification Service is suitable for various biological samples, including cells, tissues, blood, urine, etc., and is widely applied in basic research, disease diagnostics, drug development, and other fields. Based on an advanced proteomics analysis platform, MtoZ Biolabs offers the Large-Scale Protein Identification Service to help researchers efficiently extract protein information from samples.
Analysis Workflow
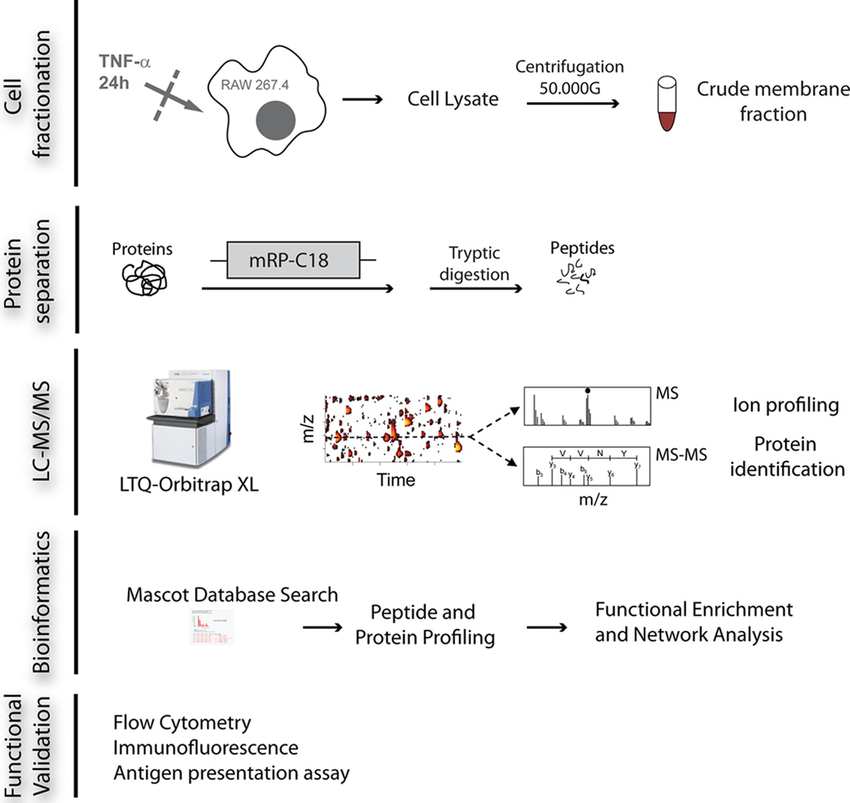
Bell C. et al. Molecular & Cellular Proteomics. 2013.
Large-Scale Protein Identification Service generally uses a bottom-up approach to analyze proteins and collects data using the DIA (Data-Independent Acquisition) mode. The main process is as follows:
1. Sample Preparation
Proteins are extracted, and may undergo concentration or impurity removal.
2. Protein Digestion
Proteins are digested using trypsin or other enzymes to generate smaller peptide fragments.
3. Liquid Chromatography Separation
The digested peptides are separated by liquid chromatography (LC).
4. Mass Spectrometry Analysis
The separated peptides are analyzed by the mass spectrometer for detection.
5. Data Analysis and Report Generation
The mass spectrometry results are matched with protein databases, and algorithms are used for peptide identification. Quantification of different proteins is performed based on the intensity of the mass spectrometry signals, and their biological significance is further analyzed through functional annotation and pathway analysis.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Large-Scale Protein Identification Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Large-Scale Protein Identification Service data, providing clients with a comprehensive data report.
Applications
1. Basic Biological Research
Perform comprehensive protein identification on large-scale samples to provide data support for fundamental research. For example, it can reveal post-transcriptional regulatory information related to gene expression and analyze how genes impact cellular functions through protein changes.
2. Disease Mechanism Research and Biomarker Discovery
Analyze proteins in pathological samples to identify potential disease biomarkers. For example, using large-scale protein identification to analyze protein differences between tumor and healthy samples can help uncover potential cancer biomarkers.
3. Drug Development
In drug screening and target validation processes, large-scale protein identification technology is used to evaluate drug targets and mechanisms. For example, by analyzing protein changes in cells or tissues, key proteins related to diseases can be identified, which can then be targeted for the development of new drugs.
FAQ
Q. How to balance the sample concentration in large-scale samples to avoid the impact of excessively high or low protein concentrations on the analysis results?
1. Sample Preprocessing and Quantification
Use precise protein quantification methods (e.g., BCA, Bradford, Lowry assays) to quantify the samples and ensure that the protein concentration is within the appropriate range for mass spectrometry analysis. The recommended protein concentration range is typically 0.1-1 mg/mL to ensure efficient peptide information capture by the mass spectrometer.
2. Dilution or Concentration
For samples with excessive protein concentration, dilution can be used to reduce the concentration, preventing overloading the mass spectrometer and causing ion suppression. For samples with low protein concentration, concentration methods (e.g., lyophilization, ultrafiltration) can be applied to increase the protein concentration, ensuring sufficient signal for analysis.
3. Stepwise Processing
For complex samples, stepwise preprocessing can be applied, such as separating proteins by molecular weight and using specific affinity capture methods (e.g., immunoprecipitation, protein enrichment) to increase the concentration of target proteins.
4. Sample Standardization
For large-scale analyses with multiple samples, standardization methods can be employed, such as normalization using internal standards, ensuring that concentration differences between samples do not affect the comparison and interpretation of results.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study used Large-Scale Protein Identification technology to systematically analyze the interaction network of LIN28A protein and identified a large number of LIN28A-related proteins from neural progenitor cells derived from human induced pluripotent stem cells. The results showed that LIN28A participated in multiple gene regulation processes, including RNA processing, translation regulation, and cell differentiation, by interacting with multiple proteins, demonstrating its important function in neural development. The study deepened the understanding of the mechanism of action of LIN28A in regulating neural progenitor cell differentiation and provided new protein interaction targets for studying stem cell differentiation and neural development.
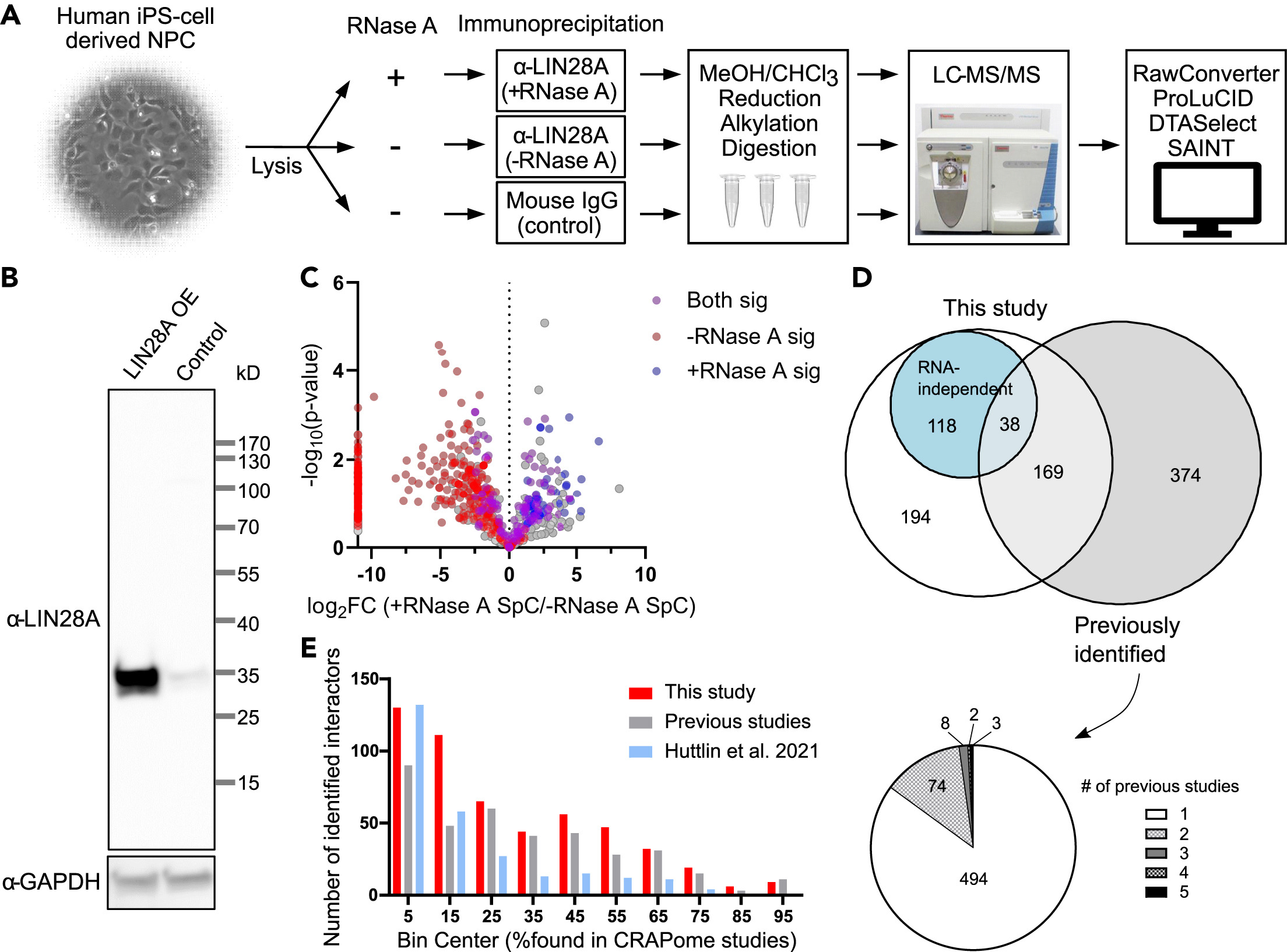
Yu N K. et al. iScience. 2021.
How to order?







