Protein Quantitation Assays Service
Proteins are the functional executors of life activities, and their concentration and distribution directly affect cellular functions and physiological processes. Therefore, protein quantification is a crucial step in biomedical research, drug development, and other fields. MtoZ Biolabs offers a variety of advanced protein quantification assays service to help clients accurately and efficiently analyze protein concentrations, providing reliable data support for both research and clinical applications.
Services at MtoZ Biolabs
The protein quantitation assays service provided by MtoZ Biolabs is based on mass spectrometry technology and multivariate data analysis to accurately quantify proteins. Mass spectrometry protein quantification enzymatically hydrolyzes protein samples into peptide fragments to form a peptide mixture; then high-performance liquid chromatography and other technologies are used to quantitate the peptides. The mixture of segments is separated into individual peptide segments; the separated peptide segments are then subjected to mass spectrometry analysis to determine the mass-to-charge ratio and intensity of each peptide segment; finally, based on the abundance of the peptide segments and the specificity of protein enzymatic hydrolysis, the protein content is calculated relative content.
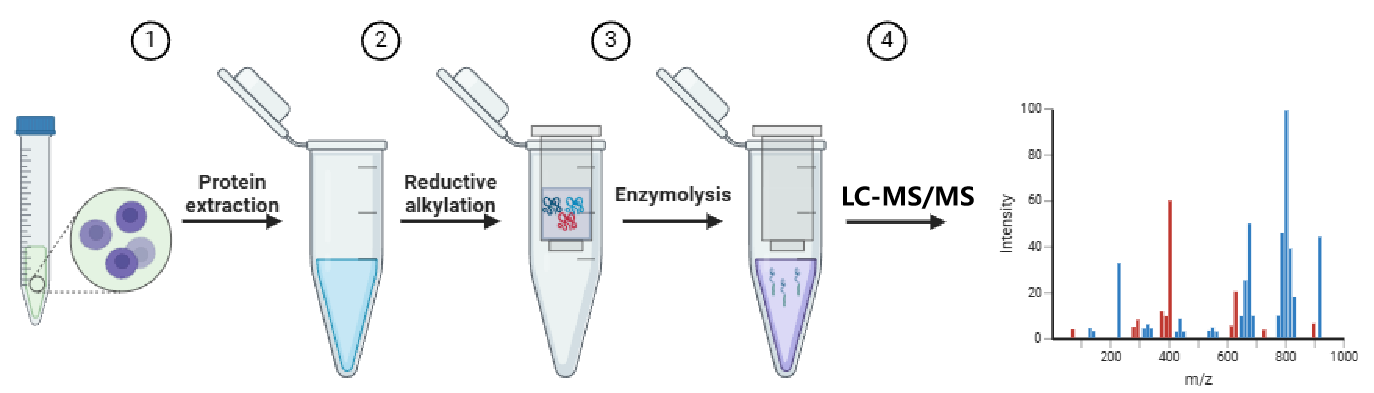
Figure 1. Schematic flow chart of mass spectrometry protein loading.
MtoZ Biolabs' mass spectrometry-based protein quantification assay service rely on non-targeted and targeted mass spectrometry quantification methods:
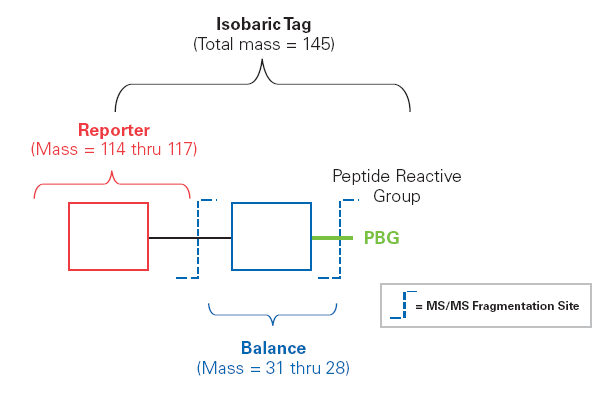
(1) Non-targeted mass spectrometry quantitative methods include: labeled quantitative methods and non-labeled quantitative methods. The labeling quantification method compares the abundance difference of proteins in labeled samples and non-labeled samples by labeling proteins in samples, such as using isotope labels, fluorescent labels, etc., thereby determining the relative content of proteins, such as using iTRAQ/TMT labeled protein quantitative analysis, Label-free quantitative methods do not use labels and directly compare the abundance differences of proteins in different samples, such as using Label-Free, DIA, and SWATH for protein quantitative analysis.
www.thermofisher.cn
Figure 2. Schematic diagram of iTRAQ/TMT labeling principle.
(2) Targeted Mass Spectrometry Quantification includes SRM/MRM and PRM. PRM allows for selective detection of target proteins or target peptides (such as peptides with post-translational modifications), enabling the quantification of target proteins/peptides. It provides high specificity and high accuracy in quantifying the target proteins, and when combined with isotope internal standards, absolute quantification can be achieved.
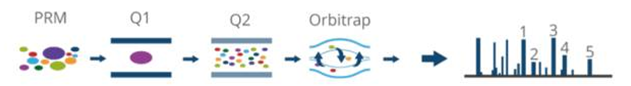
Figure 3. Schematic diagram of PRM detection principle.
Service Advantages
1. Multiple Methods Available
Choose the most suitable protein quantification assays service based on client needs to meet various experimental requirements.
2. High Accuracy and Sensitivity
MtoZ Biolabs uses Thermo's Orbitrap series mass spectrometers combined with Nano-LC systems to ensure high precision and sensitivity in data collection, guaranteeing the accuracy and reliability of quantification results.
3. Fast and Efficient
With efficient experimental processes and data handling, MtoZ Biolabs shortens the experimental cycle.
4. Professional Team
An experienced technical team provides professional support and consultation services to resolve different research issues and experimental requirements.
Sample Submission Suggestions
1. Sample Types
We accept various types of biological samples, including cells, tissues, and bodily fluids.
2. Sample Requirements
Samples should be kept intact and stable to avoid degradation or contamination.
Applications
1. Proteomics Research
Quantitative analysis of changes in protein expression levels under different conditions.
2. Disease Diagnosis and Biomarker Research
Detection of specific protein concentration changes in biological samples for disease diagnosis and biomarker research.
3. Drug Development
Evaluation of the impact of drugs on protein expression levels for drug development and efficacy assessment.
4. Food and Agriculture Testing
Detection of protein content in food and agricultural products for quality control and safety assessment.
Case Study
1. Exploring Potential Targets for Treating Neuropathic Pain Using MS-based Label-Free Protein Quantification
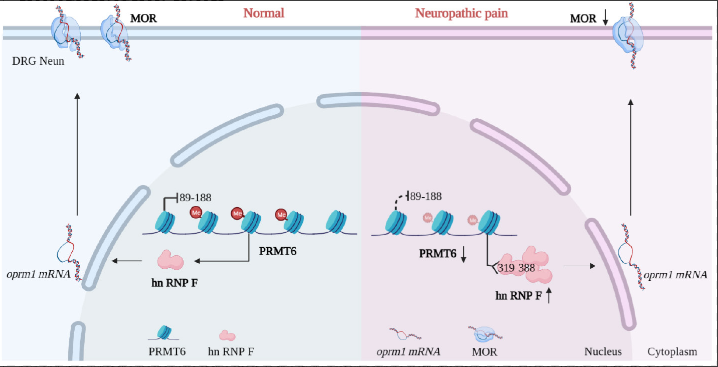
Protein arginine methyltransferase-6 participates in various biological functions, particularly RNA processing, transcription, chromatin remodeling, and endosomal trafficking. This study utilized immunohistochemistry, western blotting, immunoprecipitation, and label-free proteomics methods to investigate the expression levels of arginine methyltransferase-6 protein and its impact on neuropathic pain in models including spinal nerve injury, chronic constriction injury, and bone cancer pain. This case demonstrates how MS-based label-free protein quantification services identified arginine methyltransferase-6 as a potential therapeutic target for treating peripheral neuropathic pain, elucidating disease mechanisms and guiding treatment development.
Zhang XY et al. Neural Regeneration Research, 2024.
Figure 4. Protein arginine methyltransferase-6 regulates the expression of heterogeneous nuclear ribonucleoprotein-F.
2. Exploring the Molecular Mechanism of Inhibiting GBM Proliferation and TMZ Benefits Using TMT-based Protein Quantification
TRIM22, a ligand for the nuclear factor kappa B (NF-kB), plays a key role in the proliferation and drug sensitivity of glioblastoma multiforme (GBM). This study used cellular senescence assays, BiFC assays, and TMT-based protein quantification assays service to investigate the molecular mechanisms of the protein network between TRIM22 and NF-kB. This case demonstrates how TMT protein quantification analysis identified that the TRIM22-NT5C2 complex inhibits GBM proliferation and enhances TMZ benefits through post-translational modifications of RIG-I and modulation of the RIG-I/NF-kB/CCAR1 pathway, establishing it as a promising target for multi-target therapy in a single pathway.
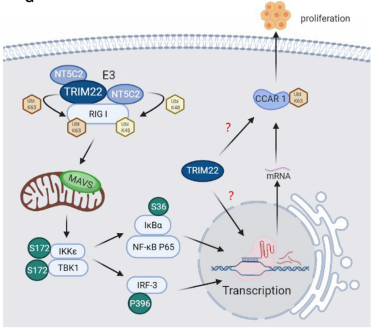
Fei, XW et al. Molecular Therap, 2022.
Figure 5. Regulation flow chart of TRIM22-NT5C2 complex.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Relative Protein Quantitative Service, MS Based
How to order?







