Protein Phosphorylation Identification Service
- Comprehensive experimental details
- Materials, instruments and methods
- High-quality spectra and visual charts
- Site-specific phosphorylation identification with detailed annotations
- Bioinformatics analysis
- Raw data files
Protein phosphorylation identification based on mass spectrometry is a key method for studying post-translational modifications of proteins, enabling accurate characterization of phosphorylation types and their site-specific distributions. Phosphorylation is a dynamic and reversible process mediated by kinases and phosphatases that add or remove phosphate groups from serine, threonine, or tyrosine residues. This modification regulates protein conformation, activity, and interactions, and plays a central role in cell proliferation, differentiation, apoptosis, metabolism, signal transduction, and disease progression.
By performing protein phosphorylation identification, researchers can map phosphorylation, analyze signaling networks, study disease mechanisms, and gain critical evidence for drug development and biomarker discovery. MtoZ Biolabs provides high-quality protein phosphorylation identification services supported by advanced LC-MS/MS platforms and specialized bioinformatics workflows, providing reliable data to support basic and applied research.
Services of MtoZ Biolabs
MtoZ Biolabs manages the entire process, from sample preparation to result interpretation, through rigorous quality control. To address the challenges of low abundance and wide dynamic range in phosphoproteins, our protein phosphorylation identification services employ enrichment methods such as TiO₂, IMAC, and antibody-based strategies for efficient capture and accurate identification. Based on sample characteristics, we select a tailored method to ensure high resolution, specificity, and interpretable results.
Analyze Workflows
1. Sample Preparation: protein extraction and enzymatic digestion with inhibitors to prevent loss of modifications.
2. Phosphopeptide Enrichment: selective capture using TiO₂, IMAC, or antibody-based methods.
3. Mass Spectrometry Detection: high-resolution LC-MS/MS data acquisition to ensure sensitivity and coverage.
4. Data Analysis: database searching and bioinformatics processing for phosphorylation site identification and functional annotation.
5. Report Delivery: complete reports including workflows, parameters, site information, spectra, and raw data.

Why Choose MtoZ Biolabs?
✅ Advanced Analysis Platform: MtoZ Biolabs established an advanced mass spectrometry platform, guaranteeing reliable, fast, and highly accurate Protein Phosphorylation Identification Service.
✅ Multiple Enrichment Strategies: TiO₂, IMAC, and antibody-based methods enhance detection of diverse samples.
✅ Tiered Detection: Differentiates mono- and multi-phosphorylated peptides, reducing ionization bias and improving precision.
✅ Expert Support: An experienced team guides the entire process, ensuring experimental design and interpretation meet project needs.
✅ One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
Sample Submission Suggestions
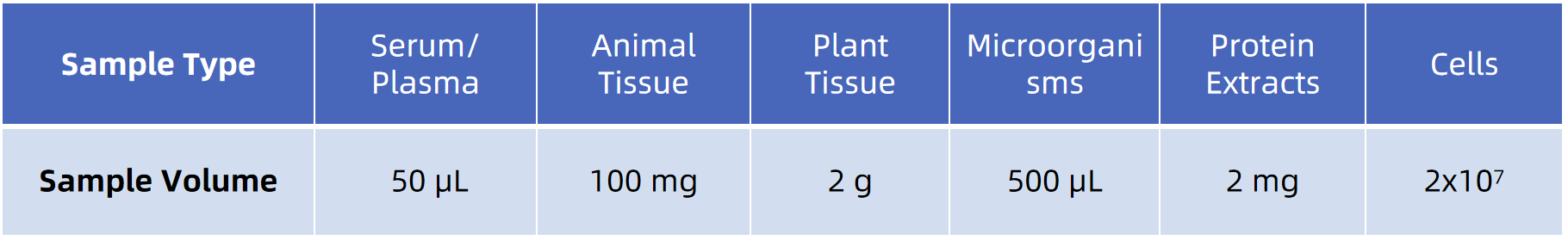
Note: To ensure reliable results, all samples should be stored at low temperatures and avoid repeated freeze-thaw cycles. For detailed submission requirements, please contact MtoZ Biolabs for a complete guide.
Applications
● Signaling Pathway Studies: Reveal downstream kinase-regulated networks.
● Disease Mechanism Research: Investigate abnormal phosphorylation events in cancer, neurodegeneration, and metabolic disorders.
● Drug Development: Assess the regulation of kinase activity and signaling pathways by candidate drugs.
● Biomarker Discovery: Identify disease-related phosphorylation sites to support precision medicine.
● Systems Biology: Construct large-scale phosphorylation maps to enable multi-omics integration.
What Can Be Included in the Report?
FAQs
Q1: Can Phosphorylation of Serine, Threonine, and Tyrosine be Identified Simultaneously?
A1: Yes. With high-resolution LC-MS/MS and optimized fragmentation strategies, MtoZ Biolabs can precisely define the phosphorylation of Ser, Thr, and Tyr residues with high confidence scores.
Q2: Can Low-abundance Phosphorylation Sites be Detected?
A2: Yes. We use enrichment strategies optimized for each sample type, such as TiO₂, IMAC, and antibody-based capture, to increase the detection efficiency of low-abundance phosphopeptides to ensure reliable results.
Q3: Can both Monophosphorylation and Polyphosphorylation be Quantified?
A3: Yes. Our layered detection strategy distinguishes between monophosphorylated and polyphosphorylated peptides, minimizing ionization bias and providing more accurate quantitative data.
MtoZ Biolabs offers comprehensive protein phosphorylation identification services with advanced instrumentation, diverse enrichment methods, and specialized bioinformatics support. Our services provide insights into signal regulation, accelerate disease research, and aid in drug development and biomarker discovery. For more details, please feel free to consult us.
Related Services
How to order?







