Parallel Reaction Monitoring (PRM) Service
Parallel reaction monitoring (PRM) is an ion monitoring technology based on high-resolution and high-precision mass spectrometry. The principle of PRM technology is similar to that of SRM/MRM, but in the development of analytical methods, PRM is more commonly used for the absolute quantification of proteins and peptides. PRM technology can detect multiple proteins at the attomole level in complex samples.
PRM is a quadrupole high-resolution mass spectrometry detection platform based on Q-Orbitrap. Unlike SRM, which performs one transition at a time, PRM performs a full scan of each transition through precursor ions, meaning that it monitors all fragments of the precursor ions simultaneously. First, PRM uses the quadrupole (Q1) to select precursor ions, with a selection range typically of m/z ≤ 2; then, the precursor ions are fragmented in the collision cell (Q2); finally, Orbitrap replaces Q3, scanning all fragmented ions with high resolution and high precision. Therefore, PRM technology not only has the quantitative analysis capability of SRM/MRM targets but also qualitative capabilities.
Analysis Workflow
1. Development of PRM Analytical Method
First, select 2 or 3 targeted peptides from the mass spectrometry library or Skyline software. The selected peptides should be unique to the target protein, easily detectable by LC-MS, without missing cleavage sites and multiple modified amino acids. Secondly, establish a standard curve based on the x-axis concentration ratio and the y-axis peak area ratio.
2. Mass Spectrometry Analysis
The hydrolyzed samples are analyzed using Thermo's latest Orbitrap Fusion Lumos mass spectrometer.
3. Data Analysis
Data analysis is performed using Skyline software.
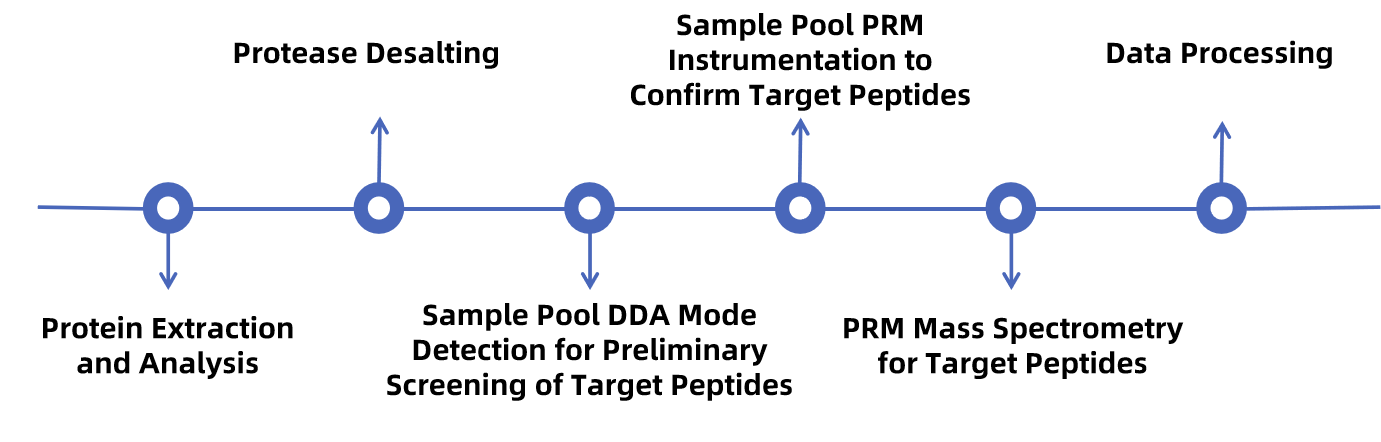
Figure 1. Workflow for PRM
Service Advantages
1. Higher Sensitivity
The mass accuracy can reach ppm levels, which better eliminates background interference and false positives than SRM/MRM, effectively improving detection limits and sensitivity in complex backgrounds.
2. Easier Development of Analytical Methods
Full scan of product ions, no need to select ion pairs, making the establishment of analytical methods easier.
3. Wider Linear Range
Order of magnitude increase to 5-6.
Sample Submission Requirements
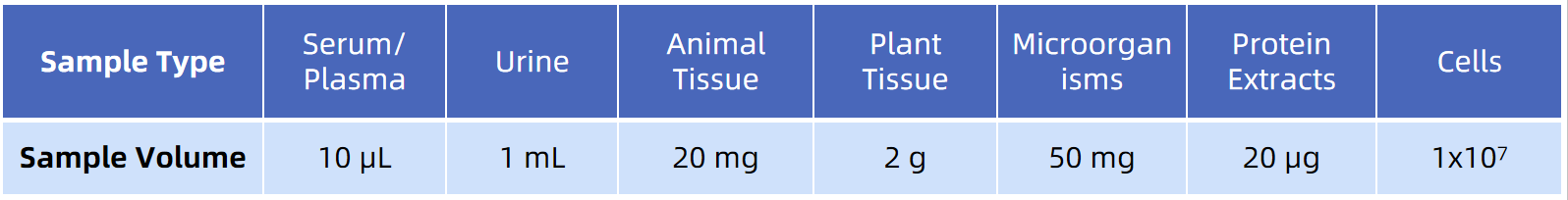
For more sample details, please consult our technical team.
Applications
1. Validation of Differential Proteins in iTRAQ
2. Validation of Label-Free Differential Proteins
3. Absolute Quantification of Peptides and Proteins
4. Quantification of Disease Markers, Establishing Diagnostic Models
5. Quantification of Phosphorylated Proteins, Methylated Proteins
6. Quantification of other Post-Translational Modified Proteins
7. Quantitative Analysis of Metabolic Pathways
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
MRM/PRM Quantitative Proteomics Service
SRM/PRM (Selected/Parallel Reaction Monitoring)-Based Targeted Validation Service
How to order?







