SRM/PRM (Selected/Parallel Reaction Monitoring)-Based Targeted Validation Service
- Validation of phosphorylation proteins identified by iTRAQ/TMT quantitative proteomics.
- Follow-up confirmation of phosphorylation sites from label-free quantitative studies.
- Absolute quantification of targeted phosphorylation peptides and proteins.
- Quantitative verification of phosphorylation biomarkers for diagnostic model development.
- Precise quantification of disease-related phosphorylation modifications.
- Quantitative analysis of phosphorylation signaling pathways and networks.
The SRM/PRM (Selected/Parallel Reaction Monitoring)-Based Targeted Validation Service is a specialized targeted mass spectrometry approach based on high-resolution, high-accuracy ion monitoring techniques, designed for precise quantification of phosphorylation events within complex biological samples. Selected Reaction Monitoring (SRM), also known as Multiple Reaction Monitoring (MRM), utilizes triple-quadrupole mass spectrometry to selectively monitor predefined precursor and fragment ion pairs, enabling highly sensitive and specific quantification. In contrast, Parallel Reaction Monitoring (PRM) employs high-resolution Orbitrap mass spectrometry, simultaneously acquiring full fragment-ion spectra of targeted precursors with ppm-level mass accuracy. This approach significantly reduces background interference and false-positive signals, enhancing sensitivity and reliability compared to SRM/MRM methods.
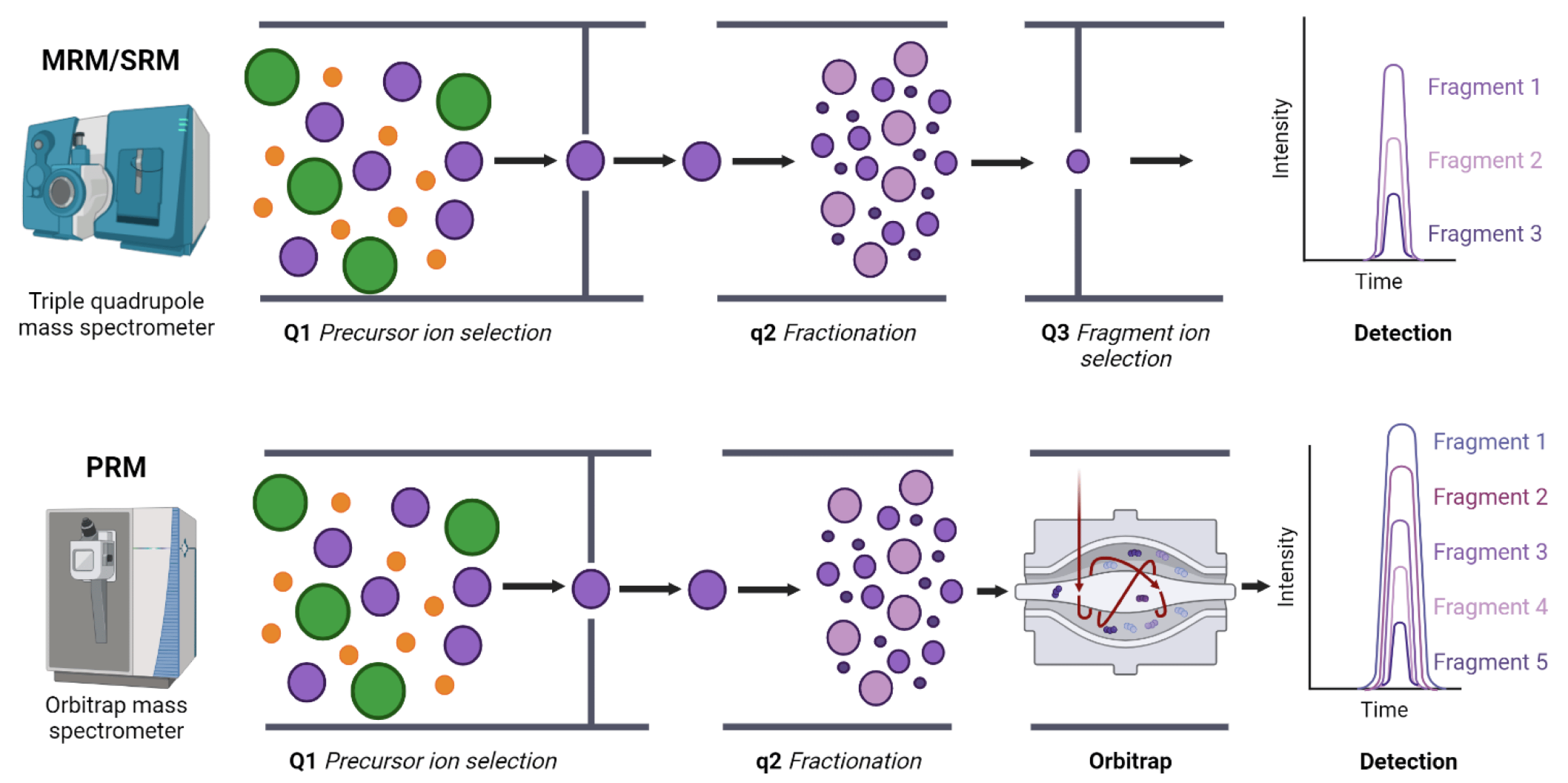
Figure 1. Multiple/Selected Reaction Monitoring (MRM/SRM) and Parallel Reaction Monitoring (PRM) Targeted Proteomics
SRM/PRM-based targeted validation is particularly effective for antibody-free validation of protein candidates identified through relative quantitative proteomics (e.g., label-free, iTRAQ, TMT, SILAC) and for targeted quantification of phosphorylated peptides. These approaches overcome limitations of traditional antibody-based assays, eliminating cross-reactivity and improving reproducibility across experimental conditions.
Service at MtoZ Biolabs
Service Advantages
✅High Sensitivity: Dual-stage ion selection effectively eliminates interfering ions, dramatically enhancing the signal-to-noise ratio and ensuring precise detection of targeted phosphopeptides.
✅High Throughput: Enables simultaneous targeted validation of hundreds of proteins in a single assay, significantly increasing research efficiency.
✅Broad Quantitative Range: Precisely identifies and quantifies low-abundance phosphorylated protein across a dynamic range spanning four orders of magnitude.
✅Absolute Quantification Capability: Enables absolute quantification of phosphorylation sites without dependence on antibody-based methods.
Applications
Case Study
Phosphoproteomic Analysis of Rice Blast Fungus
Researchers applied PRM-Based Targeted Validation to explore phosphorylation-driven infection mechanisms of the rice blast fungus, Magnaporthe oryzae. A comprehensive phosphoproteomic analysis identified 8,005 phosphosites during fungal appressorium formation, revealing critical signaling pathway rewiring. Leveraging PRM, the team validated 32 phosphoproteins as substrates of the fungal kinase Pmk1, crucial for infection. They demonstrated that Pmk1-dependent phosphorylation of regulator Vts1 was essential for rice blast disease progression. MtoZ Biolabs’ SRM/PRM (Selected/Parallel Reaction Monitoring)-Based Targeted Validation Service enables similar high-precision validation and quantification of phosphorylation sites, providing robust, antibody-free confirmation of critical signaling targets, facilitating targeted strategies for crop disease control.
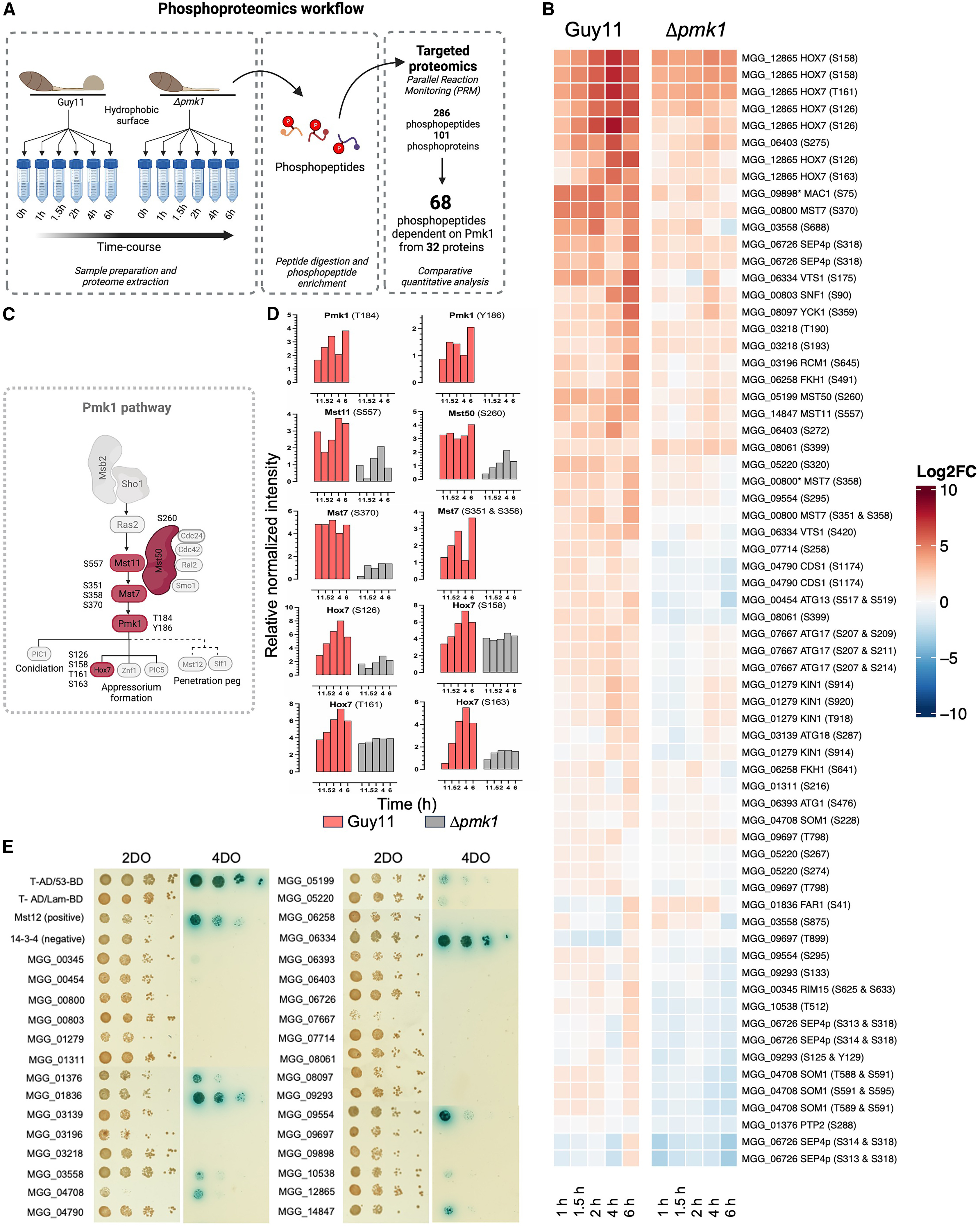
Figure 2. Quantitative Phosphoproteomics Defines 32 Putative Targets of the Pmk1 MAPK Pathway
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
MRM/PRM Quantitative Proteomics Service
How to order?







