Post-Translational Modification Site Prediction Service
- Phosphorylation site prediction
- Glycosylation site prediction
- Ubiquitination site prediction
- Mono-, di-, and tri-methylation site prediction
- Additional PTM types upon request
- Sequence motif enrichment and conservation analysis
- Kinase–substrate interaction prediction
- Cross-annotation with structural domains and functional regions
Post-translational modifications (PTMs) are fundamental mechanisms regulating protein functionality, including activity, stability, localization, and molecular interactions. These modifications—such as phosphorylation, acetylation, ubiquitination, methylation, and glycosylation—are widely involved in signaling transduction, metabolic regulation, and immune response. While high-throughput proteomics has facilitated PTM site identification, experimental validation remains costly, low-throughput, and limited in capturing dynamic changes. In this context, bioinformatics-based PTM site prediction emerges as a powerful strategy for hypothesis generation and experimental prioritization.
MtoZ Biolabs offers advanced Post-Translational Modification Site Prediction Service that integrates curated databases with state-of-the-art machine learning algorithms. Designed to support protein functional analysis and regulatory mechanism exploration, our service delivers accurate and scalable predictions across multiple modification types and biological contexts.
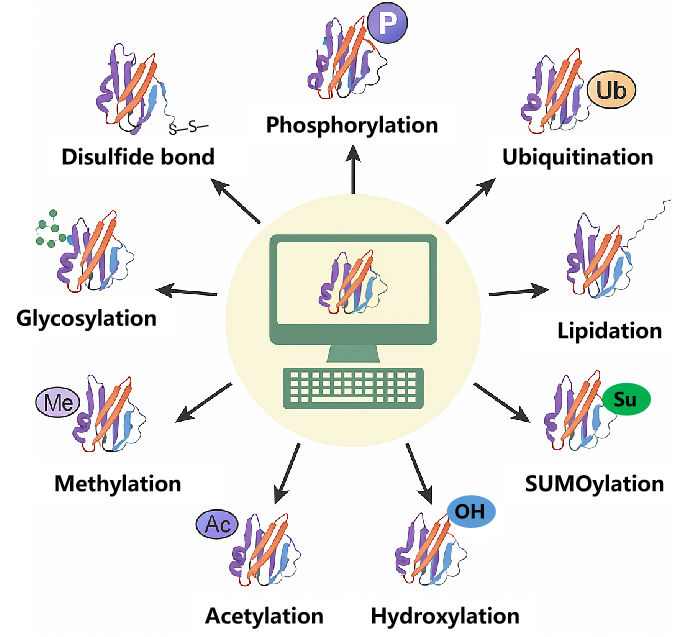
Figure 1. Overview of Common PTMs and Their Computational Prediction
Services at MtoZ Biolabs
MtoZ Biolabs’ Post-Translational Modification Site Prediction Service supports a broad range of common modification types and offers customizable predictions based on species, tissue context, or biological pathways.
Our service includes, but is not limited to, the following modification types:
Complementary analyses include:
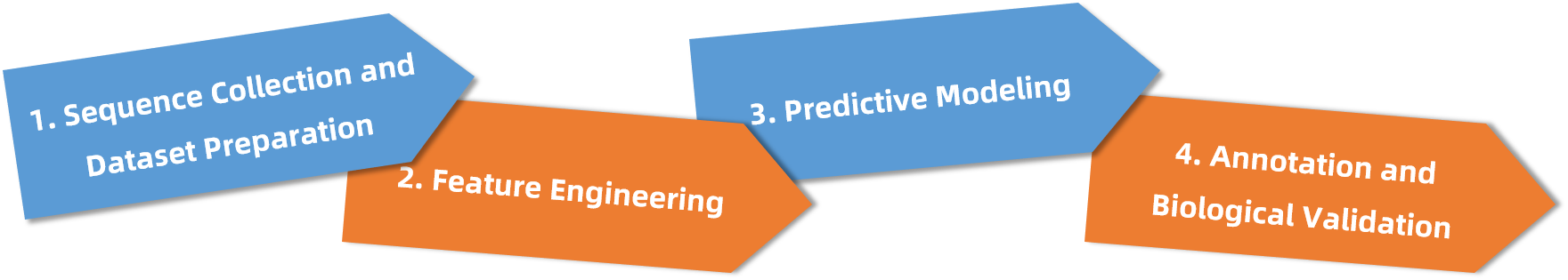
Analysis Workflow
Our bioinformatics workflow combines standardized pipelines with customized algorithmic strategies to ensure scientific rigor and biological interpretability:
1. Sequence Collection and Dataset Preparation
Target sequences (FASTA or accession IDs) are curated and used to generate high-quality training sets from reference PTM databases.
2. Feature Engineering
Multi-parametric feature matrices are constructed based on local sequence context, secondary structure, hydrophobicity, and evolutionary conservation.
3. Predictive Modeling
Machine learning models (e.g., Support Vector Machines, Random Forest) or deep neural networks are applied to generate site-level modification scores and confidence estimates.
4. Annotation and Biological Validation
Predicted sites are cross-referenced with public PTM databases and annotated for domain positioning, functional relevance, and protein–protein interaction regions (PPIRs), enhancing downstream applicability.
Why Choose MtoZ Biolabs?
MtoZ Biolabs’ service stands out for its analytical robustness and predictive accuracy:
✅ High-Coverage PTM Data Integration: Aggregated from PhosphoSitePlus, UniProt, dbPTM, and other curated databases for enhanced coverage and annotation depth.
✅ Advanced Prediction Algorithms: Supports both canonical and non-canonical motifs, with strong performance even in low-conservation regions.
✅ PPIR Localization Insights: Highlights whether modification sites are located within known or predicted interaction interfaces, aiding functional inference.
✅ High-Throughput Compatibility: Supports batch prediction of thousands of protein sequences for proteomics-scale studies.
✅ Comprehensive Annotation: Merges structural, motif-based, and pathway-level interpretations to support systems biology applications.
Applications
The Post-Translational Modification Site Prediction Service service enables deeper insights across various scientific and translational disciplines:
· Mechanistic Studies: Identify candidate regulatory sites to guide functional experiments
· Target Engineering: Inform design of PTM-null mutants or regulatory switches
· Interaction Network Mapping: Investigate PTM impact on protein interaction dynamics
· Signal Pathway Reconstruction: Aid in modeling kinase–substrate interaction networks
· Drug Discovery Support: Predict functional sites that may influence drug binding or efficacy
What Could be Included in the Report?
1. PTM prediction table (site number, residue, modification type, confidence score, PPIR status)
2. Motif enrichment summary and visualization charts
3. 3D site mapping (if structural models are available)
4. Annotated report including algorithms, tools, and database references
5. Input and intermediate files for traceability and reproducibility
As part of our integrated proteomics solutions, MtoZ Biolabs’ Post-Translational Modification Site Prediction Service complements our extensive suite of experimental PTM services, including enrichment-based quantification and functional annotation of phosphorylation, glycosylation, acetylation, and ubiquitination. Let our expertise accelerate your discovery—reach out to MtoZ Biolabs for a customized consultation today.
Related Services
PRM-based Post-translational Modification Site Analysis Service
Post-translational Modifications (PTMs) Service
Post-Translational Modification Crosstalk Prediction Service
How to order?







