Peptidomics-Based Antigen Discovery and Prediction Service
Peptidomics-based antigen discovery and prediction is a technique that uses high-resolution mass spectrometry to identify and analyze peptides that bind to immune receptors, thereby discovering potential immune epitopes or antigens. This service employs advanced techniques for extracting, purifying, and enriching peptides, combined with LC-MS/MS technology, to accurately identify peptides that bind to immune receptors. By analyzing peptide sequences, abundance, modification states, and other factors, it can identify potential immune epitopes and new antigens, providing data support for immunotherapy and vaccine development.
The peptidomics-based antigen discovery and prediction service is widely applied in tumor immunology, vaccine development, infection immunity, immune monitoring, and other fields. By screening and analyzing peptides interacting with the immune system, researchers can discover new immune epitopes, evaluate the immunogenicity of peptides, and provide foundational data for the development of immune targets. This service offers an important tool for immunology research, advancing personalized immunotherapy and the development of precision vaccines.
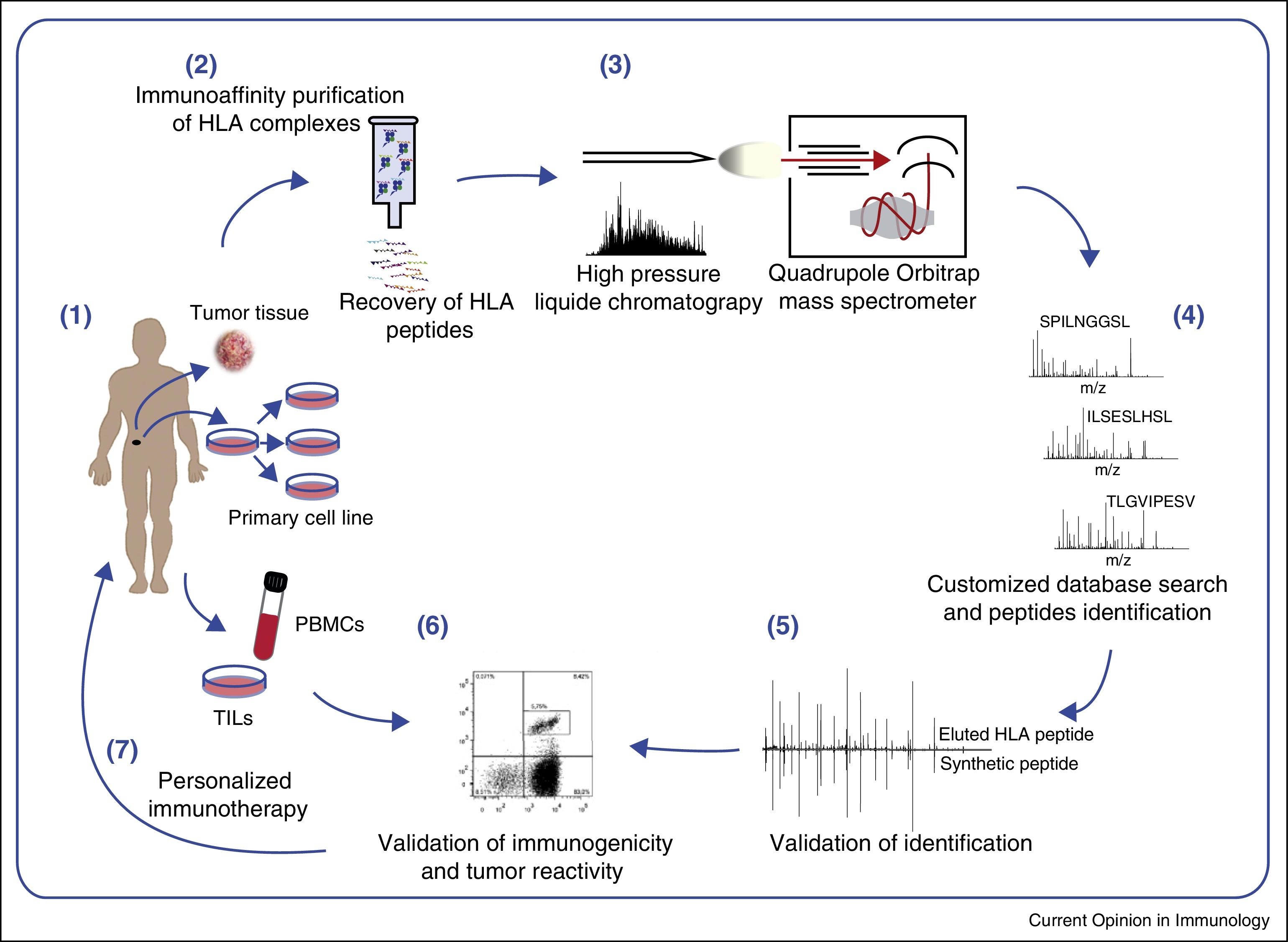
Bassani-Sternberg, M. et al. Current Opinion in Immunology, 2016.
Figure 1. Antigen Discovery by MS-Based Immunopeptidomics for Personalized Cancer Immunotherapy.
Services at MtoZ Biolabs
Based on high-resolution mass spectrometry platforms, MtoZ Biolabs offers the peptidomics-based antigen discovery and prediction service which utilizes mass spectrometry to comprehensively analyze peptide libraries, discovering and predicting antigens that bind to immune receptors. The service involves extracting and purifying peptides related to the immune system, followed by precise peptide analysis using mass spectrometry. Clients can obtain data on peptide sequences, abundance distribution, modification states, and other key information, providing valuable support for further research. The services offered by MtoZ Biolabs include, but are not limited to, the following:
1. Epitope Mapping Analysis
Analyze peptides bound to immune receptors and construct epitope maps to reveal the presentation characteristics of peptides in different immune contexts, providing data support for immune research.
2. Peptide-Antigen Interaction Studies
Study the interactions between peptides and antigens or immune receptors, assess their binding affinity and immune activation potential, and provide foundational data for immune target identification.
3. Validation Studies
Perform experimental validation of selected peptides to confirm their specificity and biological efficacy with immune receptors, ensuring the reliability and practical application of the data.
4. Epitope Prediction
Utilize bioinformatics tools and algorithms to predict potential T-cell or B-cell epitopes, assisting in the identification of key peptides related to immune responses.
Analysis Workflow
1. Sample Preparation
Using advanced techniques, extracted peptides are enriched and selected based on their affinity to immune receptors, ensuring high specificity for further analysis.
2. Peptide Enrichment and Screening
Immunoaffinity purification techniques are employed to enrich the extracted peptides, selecting those with high affinity to immune receptors for subsequent analysis.
3. Mass Spectrometry Analysis
High-resolution mass spectrometry platforms are used for liquid chromatography-mass spectrometry (LC-MS) analysis of the enriched peptides, providing detailed sequence, abundance, and modification data.
4. Data Analysis and Functional Validation
Immunogenicity analysis is performed using mass spectrometry data to predict potential immune epitopes. The selected epitopes are then validated, and their potential roles in immune responses are assessed, ensuring the reliability of the experimental results.
Service Advantages
1. High-Throughput Screening Capability
Through high-throughput screening technology, we can quickly analyze a large number of immune peptides, significantly improving data collection efficiency and expanding the scope of research.
2. Reliable Data Support
Utilizing mass spectrometry and advanced extraction and purification techniques, we provide high-quality data support, ensuring the accuracy and reliability of peptide identification and functional validation.
3. Customized Service
Based on the specific research needs of our clients, we offer tailored experimental plans, providing personalized antigen discovery and epitope prediction services.
4. One-Stop Service Workflow
We offer a complete service workflow from sample preparation, peptide enrichment, and mass spectrometry analysis to data interpretation, streamlining experimental operations and improving analysis efficiency.
Applications
1. Tumor Neoantigen Identification
Through peptidomics analysis of immune peptides in tumor tissues, we identify and predict potential neoantigens, providing target support for tumor immunotherapy.
2. Vaccine Epitope Screening
By screening peptides that bind to immune receptors, we help design more immunogenic vaccine epitopes, enhancing the effectiveness of vaccines.
3. Antigen Recognition
By analyzing peptides from different sources, we identify the differences between self and allo antigens, providing data support for transplant immune responses and immune rejection.
4. Immune Evasion Mechanism Research
Using peptidomics technology, we uncover immune evasion antigens in tumors or infectious diseases, providing foundational data for the development of new immunotherapy strategies.
FAQ
Q1: Does the Service Support Large-Scale High-Throughput Analysis?
A1: Yes, we use high-throughput screening technology to quickly analyze peptides from large numbers of samples, providing efficient immune epitope screening and antigen discovery services.
Q2: Can Post-Translationally Modified Peptides Be Identified?
A2: Yes, we support the identification and analysis of post-translationally modified peptides, such as phosphorylation, acetylation, and others. If needed, please contact us in advance. For specific modifications, we will optimize mass spectrometry methods to ensure precise identification of modification sites.
How to order?







