Nuclear Proteomics Services
The nucleus is the command center of eukaryotic cells, orchestrating critical processes such as DNA replication, transcriptional regulation, RNA processing, and chromatin remodeling. Proteins residing in the nucleus are intricately involved in maintaining genomic stability and mediating dynamic regulatory networks. Systematic profiling of the nuclear proteome provides deep insights into cell cycle regulation, transcription factor activity, epigenetic modifications, and nuclear signaling pathways in both physiological and disease contexts.
MtoZ Biolabs offers high-resolution Nuclear Proteomics Services designed to uncover the complexity of nuclear protein landscapes and their functional dynamics. Our integrated service platform combines advanced subcellular fractionation, state-of-the-art mass spectrometry, and deep bioinformatics analysis to support comprehensive studies of nuclear structure and function under various biological conditions.
Services at MtoZ Biolabs
Built on Orbitrap-class high-resolution MS platforms and optimized nuclear isolation protocols, our Nuclear Proteomics Services support both targeted and global exploration of nuclear proteomes:
· Global Nuclear Protein Identification
Comprehensive profiling of nuclear proteins using LC-MS/MS, including transcription regulators, RNA-binding proteins, chromatin-associated factors, and structural proteins
· Quantitative Nuclear Proteomics (Label-Free, TMT, iTRAQ)
Quantitative analysis of nuclear proteins across biological conditions using label-free or isobaric tagging methods, suitable for limited sample input or multiplexed experimental designs.
· Post-Translational Modification (PTM) Analysis
Sensitive identification of key regulatory modifications—such as phosphorylation, acetylation, and ubiquitination—revealing dynamic control mechanisms in nuclear signaling and stress responses.
· Sub-Nuclear Compartment Proteomics
Targeted analysis of nucleolus, nuclear membrane, chromatin, and other sub-nuclear domains to resolve spatially distinct proteome signatures and region-specific functions.
Analysis Workflow
MtoZ Biolabs provides a standardized and quality-controlled workflow to ensure high reproducibility, sensitivity, and biological relevance across nuclear proteomics projects:
1. Sample Preparation and Nuclear Isolation
Customized lysis methods (e.g., sonication, homogenization, or powder resuspension) are used to disrupt cells, followed by density gradient centrifugation for high-purity nuclear extraction and quality validation.
2. Nuclear Protein Extraction and Quantification
Hypotonic lysis and protein precipitation techniques (e.g., TCA/acetone or phenol extraction) enable efficient recovery of nuclear proteins for downstream quantification.
3. Proteolytic Digestion and Preprocessing
Proteins are digested with trypsin, followed by desalting and cleanup steps to enhance MS performance and detection sensitivity.
4. LC-MS/MS Acquisition (DDA/DIA)
High-resolution MS analysis is performed using DDA or DIA modes, compatible with both label-free and isobaric quantification strategies.
5. Bioinformatics and Functional Annotation
Comprehensive data analysis includes protein identification, quantification, GO/KEGG enrichment, PTM site localization, subcellular annotation, and interaction network reconstruction.
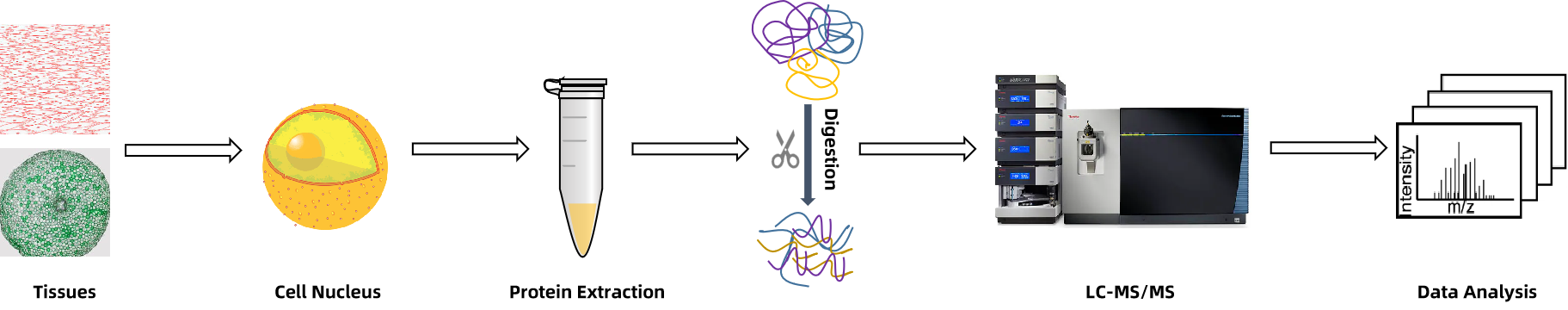
Figure 1. Nuclear Proteomics Workflow
Why Choose MtoZ Biolabs?
✅ Cross-Species Compatibility
Our workflows are optimized for nuclear proteome analysis across a variety of biological sources, including animal, plant, and microbial systems.
✅ High Nuclear Purity
Rigorous fractionation protocols ensure effective separation of nuclear from cytoplasmic proteins, enhancing proteome specificity and data quality.
✅ Flexible Acquisition Strategies
DDA, DIA, and SWATH acquisition modes are supported, enabling deeper proteome coverage and broader quantification ranges.
✅ PTM Enrichment Support
Compatible with enrichment protocols for various PTMs, including phosphorylation, acetylation, and ubiquitination, to support signaling-related research.
✅ Integrated Bioinformatics Interpretation
From database annotation to pathway enrichment and network analysis, we deliver biologically meaningful insights to support hypothesis generation and discovery.
Applications
Our Nuclear Proteomics Services can be applied to diverse areas of life science and translational research:
Fundamental Research: Dissect nuclear mechanisms underlying gene regulation, chromatin organization, RNA maturation, and spatial compartmentalization through proteome-wide expression and PTM analysis.
Disease Mechanism and Biomarker Discovery: Identify disease-associated nuclear proteins and modification events relevant to cancer, neurodegeneration, and immune disorders; uncover potential therapeutic targets such as transcription factors or DNA repair proteins.
Drug Mechanism of Action Studies: Evaluate drug-induced nuclear signaling changes by tracking alterations in nuclear proteomes and protein interaction networks (e.g., p53, NF-κB, STAT), supporting target validation and mode-of-action studies.
Plant and Agricultural Research: Investigate nuclear responses to biotic and abiotic stress (e.g., drought, salinity, pathogens), and identify key transcriptional or epigenetic regulators involved in development, adaptation, and breeding.
FAQ
Q1: Can you perform compartment-specific analysis of nucleolar, chromatin, or nuclear membrane proteins?
Yes. MtoZ Biolabs offers specialized sub-nuclear proteomics workflows. For example, nucleoli can be isolated using non-ionic lysis and ultracentrifugation, allowing for in-depth analysis of ribosome assembly factors, RNA processing proteins, and chromatin regulators—ideal for investigating RNA metabolism and nuclear structural remodeling.
If you're seeking to decode the regulatory architecture and signal integration within the nucleus, MtoZ Biolabs’ Nuclear Proteomics Services provide the precision tools and analytical depth you need. Contact us today to discuss your project—we offer customized technical solutions and full-process support to advance your research with confidence.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
Related Services
How to order?







