Mitochondrial Protein Acetylation Analysis Service
- Controlled Enrichment Efficiency: Antibody lot validation with post-enrichment acetyl-peptide ratio reports.
- Accurate Site Localization: High-resolution site-level mapping ensures precise modification assignment.
- Deep Coverage: Multi-step peptide fractionation enables detection of hundreds to thousands of acetylation sites.
- Flexible Quantification Options: Compatible with LFQ, DIA, or TMT strategies based on experimental design.
- One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
- High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all mitochondrial protein acetylation analysis service data, providing clients with a comprehensive data report.
Protein acetylation is a widespread post-translational modification (PTM) that plays a regulatory role across multiple functional modules within the cell. In recent years, mitochondrial protein acetylation has emerged as a highly dynamic PTM at the intersection of metabolomics, proteomics, and disease mechanism research. Mass spectrometry-based proteomic studies have revealed that more than 65% of mitochondrial proteins carry lysine acetylation sites, encompassing key pathways such as the tricarboxylic acid (TCA) cycle, fatty acid β-oxidation, the electron transport chain, and antioxidant defense systems. Unlike classical histone acetylation, most mitochondrial acetylation events occur via non-enzymatic mechanisms, driven by elevated levels of mitochondrial acetyl-CoA and the alkaline environment within the organelle.
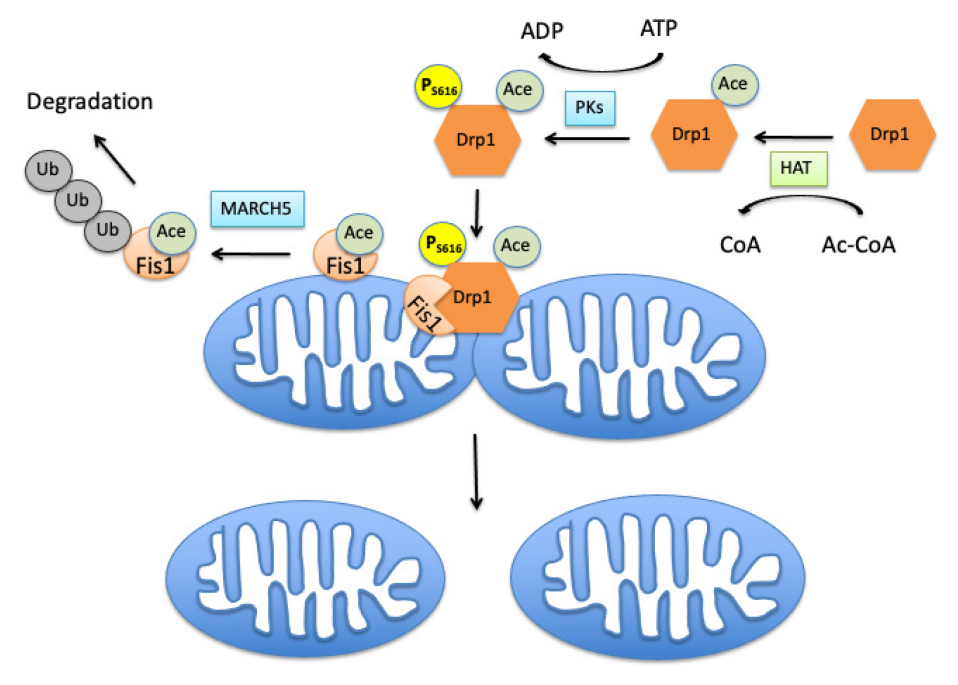
Waddell, J. et al. Cells. 2021.
Figure 1. Acetylation of Mitochondrial Fission Proteins
Key regulators of mitochondrial protein acetylation include NAD⁺-dependent deacetylases such as SIRT3, SIRT4, and SIRT5, with SIRT3 being the most extensively studied. Loss-of-function models for SIRT3 exhibit mitochondrial dysfunction, exacerbated oxidative stress, and metabolic imbalance, underscoring its central role in mitochondrial homeostasis. A growing body of research has linked aberrant mitochondrial acetylation to a wide range of diseases, including diabetes, obesity, non-alcoholic fatty liver disease (NAFLD), cardiovascular diseases, neurodegenerative disorders, and cancer. For instance, impaired SIRT3-mediated deacetylation in cardiomyocytes is considered a critical upstream event in the development of mitochondrial dysfunction, cardiac hypertrophy, and arrhythmias. Systematic mapping of the mitochondrial acetylome and comparative analysis of acetylation patterns across treatment groups or disease models have become essential strategies in uncovering disease mechanisms and identifying therapeutic targets.
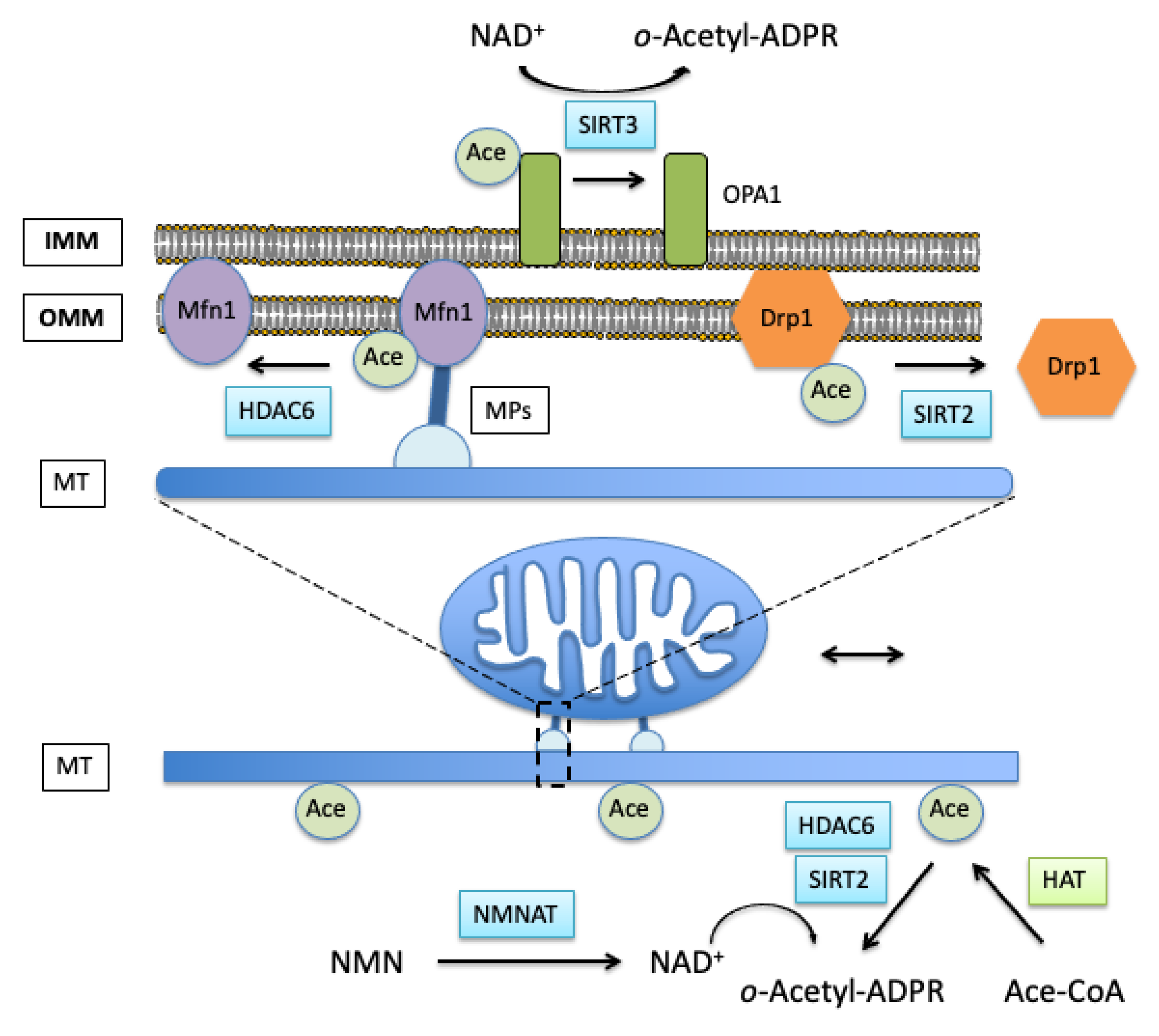
Waddell, J. et al. Cells. 2021.
Figure 2. Acetylation Effects on Mitochondrial Interaction with Microtubules and Related Fusion/Fission Proteins
Service at MtoZ Biolabs
As a CRO with long-standing expertise in proteomics and post-translational modification (PTM) research, MtoZ Biolabs offers a specialized mitochondrial protein acetylation analysis service that built on our high-purity mitochondrial isolation workflows, acetyl-lysine-specific peptide enrichment strategies, and advanced Orbitrap-based high-resolution mass spectrometry platforms. This service is designed for comprehensive identification, quantification, and pathway annotation of mitochondrial acetylation sites.
We deliver an end-to-end solution covering sample preparation, site-specific acetylation mapping, quantitative analysis, functional enrichment, and biological interpretation. The service is compatible with a wide range of experimental applications, including:
1. SIRT3/SIRT5 signaling studies
2. Glucose and lipid metabolism disorder models
3. Tumor metabolic reprogramming analysis
4. Antioxidant and redox pathway investigations
By enabling high-resolution profiling of acetylation dynamics, our platform supports advanced research in metabolic diseases, cancer biology, aging mechanisms, and therapeutic intervention strategies.
Analysis Workflow
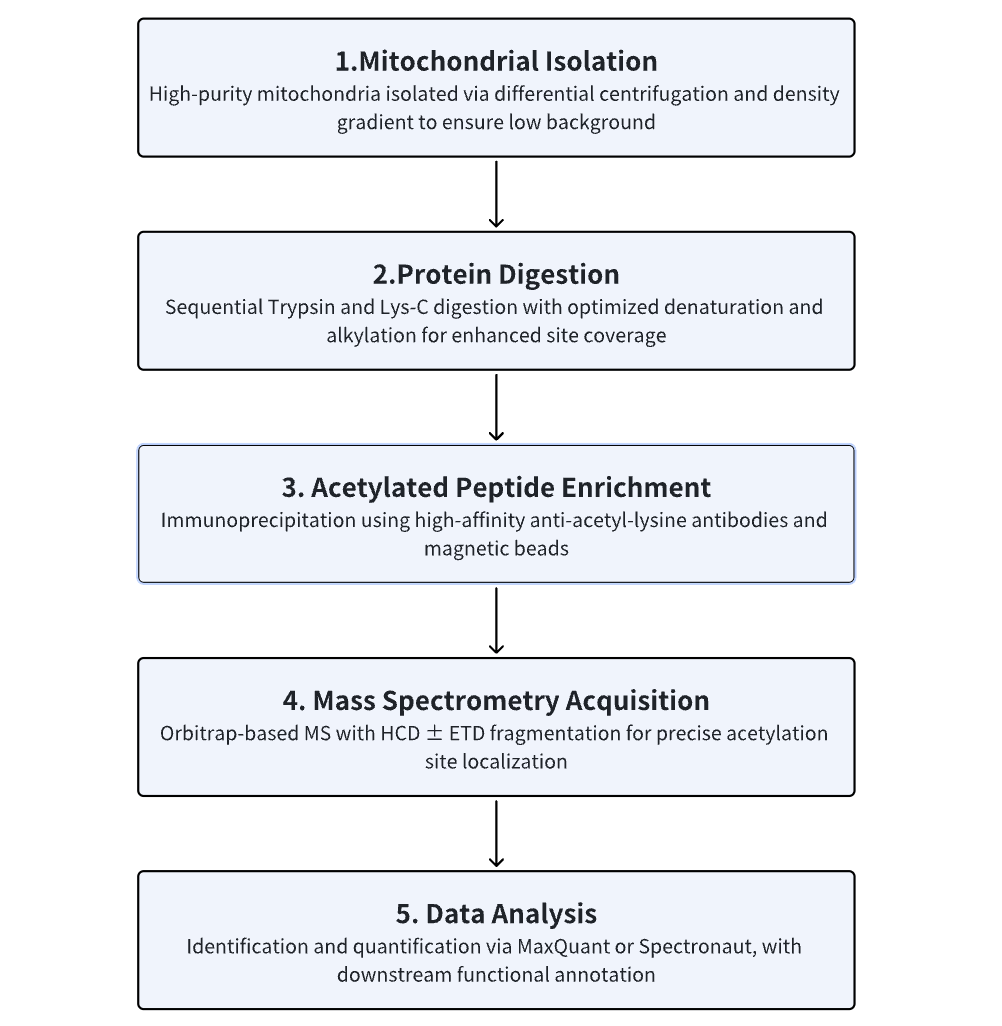
Why Choose MtoZ Biolabs
Applications
1. Metabolic Pathway Regulation
Investigate how acetylation modulates enzyme activity in the TCA cycle, fatty acid oxidation, and oxidative phosphorylation.
2. Sirtuin-Mediated Deacetylation Studies
Profile substrates and targets of mitochondrial deacetylases such as SIRT3 and SIRT5.
3. Modification Response to Treatment or Stress
Compare acetylation levels before and after drug treatment or environmental stress.
4. Cancer Metabolic Reprogramming
Analyze how acetylation shapes mitochondrial adaptation and energy metabolism in tumors.
5. Aging and Mitochondrial Dysfunction
Explore the role of acetylation imbalance in aging and neurodegenerative disorders.
6. Integrated Multi-Omics Pathway Modeling
Combine with phosphorylation, lactylation, metabolomics, or transcriptomics to reconstruct regulatory networks.
Sample Submission Suggestions
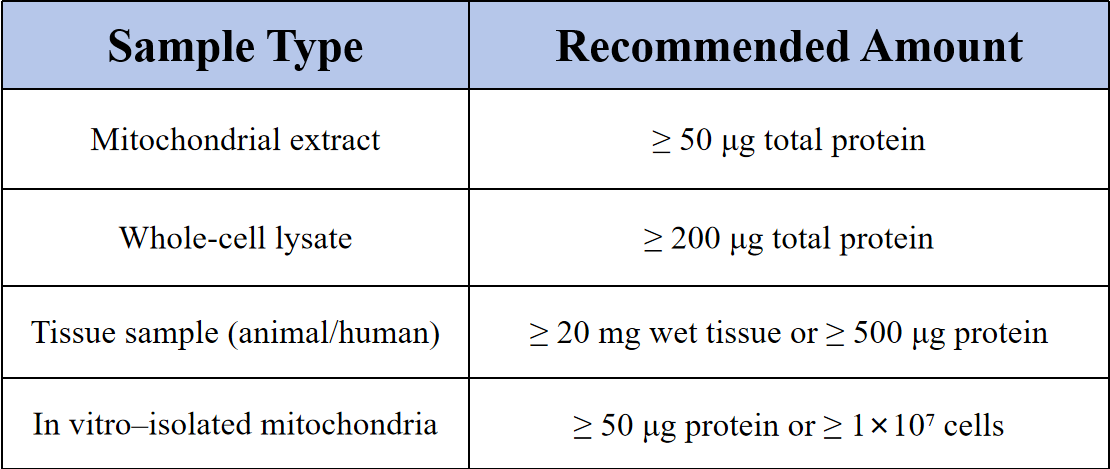
Notes:
1. Avoid using buffers containing SDS, EDTA, or high concentrations of glycerol, as these may inhibit enzymatic digestion or interfere with mass spectrometry. If non-standard buffers are used, please specify their composition in advance.
2. The use of deacetylase inhibitors is not recommended, as they may interfere with antibody-based enrichment during sample processing.
If you have any questions about sample preparation, please contact us for a detailed submission guide and technical recommendations.
Deliverables
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
How to order?







