Chloroplast Proteomics Services
Chloroplasts are specialized semi-autonomous organelles in plants, essential for photosynthetic carbon fixation and carbohydrate synthesis. Beyond this primary role, they serve as central hubs for the biosynthesis of amino acids, fatty acids, and terpenoids, and participate in the reduction of nitrate and sulfate. Due to their multifunctionality, chloroplasts are deeply embedded within the broader plant metabolic network. Notably, chloroplast proteins comprise approximately 10%–25% of total cellular proteins and exhibit distinct compartmentalization and localization, forming functionally specialized sub-proteomes.
MtoZ Biolabs offers professional Chloroplast Proteomics Services that encompass the complete workflow from chloroplast isolation and subcompartment protein extraction to high-resolution mass spectrometry and functional annotation. Our platform supports systematic studies of photosynthesis-related proteins, membrane proteins, post-translationally modified proteins, and suborganellar protein networks, providing reliable support for research in plant molecular biology and functional genomics.
Services at MtoZ Biolabs
MtoZ Biolabs’ Chloroplast Proteomics Services integrate advanced chloroplast isolation techniques with state-of-the-art mass spectrometry and deep proteomic expertise. Our services cover the following key areas:
· Chloroplast Membrane Proteome Profiling
In-depth analysis of proteins from the outer and inner envelope membranes, including transporters, lipid biosynthesis enzymes, signaling proteins, and components of nuclear-encoded protein import machinery.
· Stromal Protein Characterization
Investigation of stromal proteins involved in chloroplast genome expression, proteolytic regulation, carbon assimilation, and their associated translocation pathways.
· Thylakoid Membrane and Lumen Proteomics
Comprehensive profiling of photosystem I and II, cytochrome b6f complexes, ATP synthase, and lumen-resident low-abundance regulatory proteins involved in light energy conversion and redox signaling.
· Subcompartment-Specific Protein Network Analysis
Using region-specific proteomic strategies, we map spatially resolved protein interactions to model chloroplast sub-proteome organization and uncover metabolic regulation at the microscale.
Analysis Workflow
1. Sample Preparation and Organelle Isolation
High-purity intact chloroplasts are isolated from plant tissues using Percoll density gradient centrifugation, ensuring preservation of suborganellar structures.
2. Subcompartment Separation and Protein Extraction
Envelope membranes, stroma, and thylakoid fractions are individually separated and processed using optimized extraction buffers compatible with membrane and low-abundance proteins.
3. Proteolytic Digestion and Optional PTM Enrichment
Protein samples undergo enzymatic digestion (e.g., trypsin), and optional enrichment protocols (e.g., for phosphopeptides, propionylated peptides) are applied for targeted PTM analysis.
4. Mass Spectrometry Acquisition (LC-MS/MS)
Samples are analyzed on high-resolution Orbitrap or Q-TOF platforms using advanced acquisition modes such as DDA, PRM, and SWATH to enhance coverage and quantification of low-abundance proteins.
5. Bioinformatics and Functional Interpretation
Protein identification and functional annotation are performed using integrated databases (SUBA, PPDB, UniProt) and subcellular prediction tools, followed by network construction and pathway enrichment analysis.
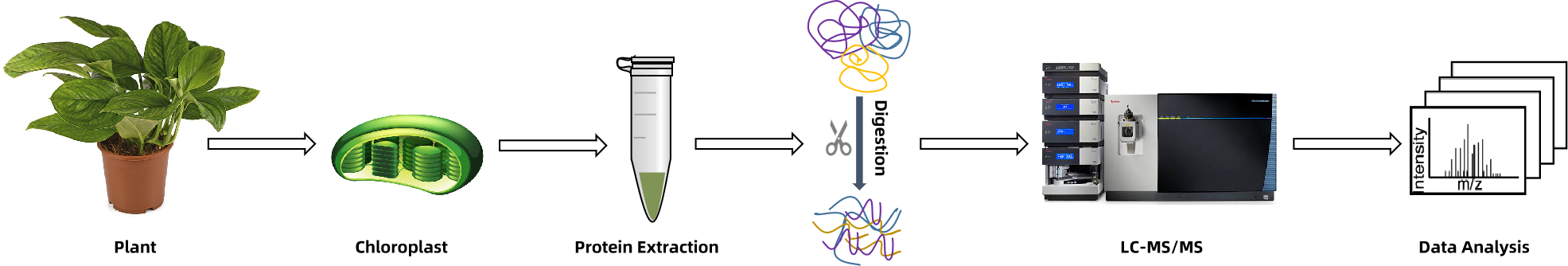
Figure 1. Chloroplast Proteomics Workflow
Why Choose MtoZ Biolabs?
✅ High-Resolution Mass Spectrometry Platforms
Powered by Orbitrap and Q-TOF systems, supporting DDA, PRM, and SWATH modes to enhance detection depth and quantification precision for low-abundance chloroplast proteins.
✅ High-Precision Sub-Organelle Fractionation
Accurate separation of chloroplast substructures—envelope, stroma, thylakoid membrane, and lumen—enables detailed sub-proteome mapping.
✅ High-Sensitivity PTM Profiling
Comprehensive analysis of post-translational modifications, including phosphorylation and acetylation, reveals key regulatory mechanisms in chloroplast function.
✅ Comprehensive Protein Annotation Pipeline
Integrated analysis using SUBA, PPDB, and UniProt databases combined with localization and enrichment tools ensures accurate functional insights.
✅ Broadly Customizable Workflow
Flexible service packages from sample prep to data analysis, tailored for stress response studies, mutant profiling, and photosynthesis research.
Applications
Our Chloroplast Proteomics Services support a wide range of plant research applications:
· Photosynthetic Complex Analysis: Identification and quantification of core proteins from PSI, PSII, Cytb6f, and ATP synthase complexes to investigate the regulation of light-dependent reactions.
· Stress-Induced Protein Expression Dynamics: Comparative analysis of chloroplast proteomes under abiotic stresses (e.g., drought, salinity, cold) to identify responsive proteins and adaptive mechanisms.
· Membrane Protein Localization and Functional Validation: Subcellular localization of envelope and thylakoid membrane proteins to refine compartment-specific proteome models.
· Mutant and Transgenic Line Support: Proteomic profiling of mutant or transgenic plants to reveal pathway-level effects and regulatory targets of gene perturbation.
For researchers aiming to uncover the roles of chloroplast proteins in plant development, stress adaptation, and metabolic regulation, MtoZ Biolabs offers reliable and in-depth Chloroplast Proteomics Services. We also provide a comprehensive suite of Post-translational Modification Proteomics Services—including phosphorylation, acetylation, propionylation, and succinylation—to support diverse research needs in plant proteomics. Reach out to our team to explore tailored solutions and technical details for your next project.
FAQ
Q1: How does chloroplast proteomics differ from whole-plant proteomics?
Chloroplast proteomics focuses on organelle-enriched samples, enabling specific and sensitive detection of proteins involved in photosynthesis and plastid-specific metabolism. In contrast, whole-plant proteomics is challenged by high background and low specificity. Our chloroplast enrichment workflow enhances signal quality and depth of analysis, making it ideal for mechanistic investigations and differential expression studies.
Q2: How can I quantify low-abundance chloroplast proteins?
Using high-sensitivity MS platforms and targeted acquisition methods like PRM and SWATH, we achieve accurate quantification of low-abundance proteins, including regulatory factors and signaling components often missed in untargeted analyses.
What Could be Included in the Report?
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
Related Services
Chloroplast Isolation and Chloroplast Protein Purification Service
How to order?







