Methylation Proteomics Service
- Epigenetic Mechanism Studies: Reveal the relationship between methylation and chromatin remodeling or transcriptional regulation.
- Cancer and Disease Mechanism Research: Identify disease-associated methylation targets and regulatory networks.
- Signal Transduction and Transcription Factor Regulation Analysis: Elucidate the role of methylation in cellular signaling pathways.
- Drug Target Screening and Mechanism Validation: Evaluate the impact of candidate compounds on key methylation sites.
- Systems Proteomics Research: Integrate methylation data with other PTMs to construct a multidimensional regulatory landscape.
Methylation Proteomics Service refers to a specialized service that utilizes high-resolution mass spectrometry to comprehensively identify, quantify, and functionally annotate methylation sites on proteins. This service not only detects the methylation status of specific proteins but also deciphers the dynamic changes of methylation modifications under various physiological or pathological conditions, thereby uncovering their precise roles in biological regulation.
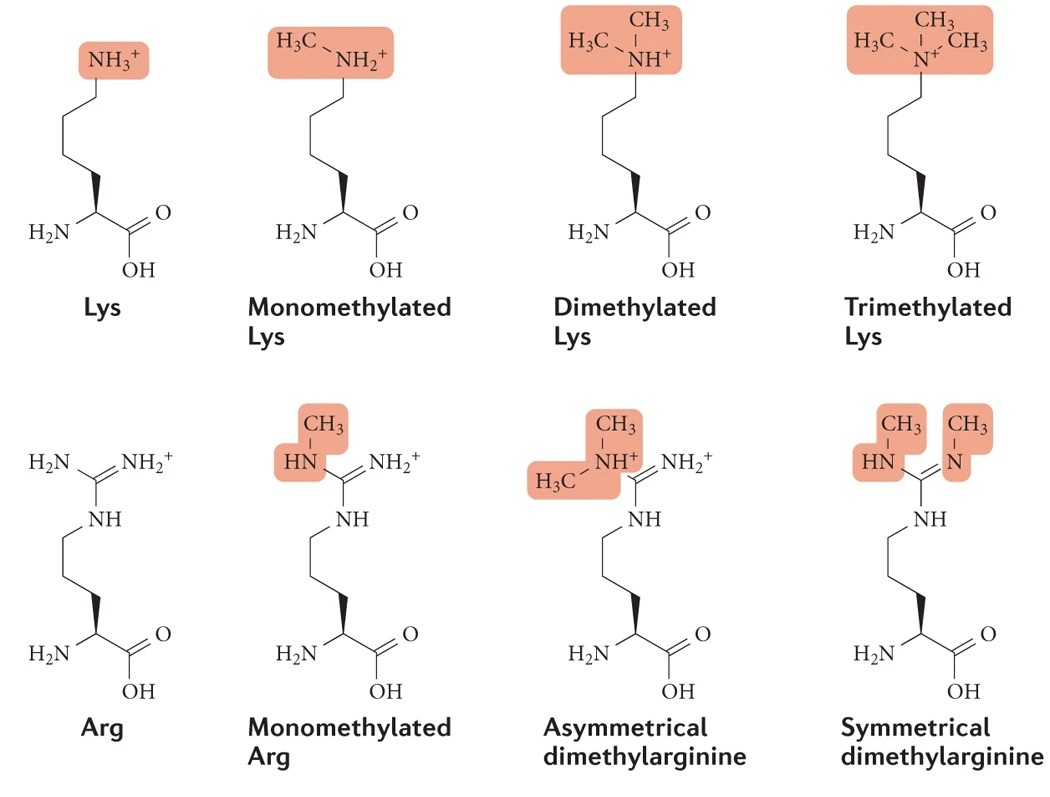
Biggar K. Li SC. Nat Rev Mol Cell Biol. 2015.
Protein methylation refers to the covalent modification of specific amino acid residues in proteins by the addition of methyl groups, most commonly occurring on lysine (Lys) and arginine (Arg) residues. This modification can regulate protein conformation, subcellular localization, stability, and interactions with nucleic acids or other proteins by adding one or more methyl groups. Methylation plays a crucial role not only in epigenetic regulation but also in key biological processes such as signal transduction, transcriptional control, RNA processing, DNA repair, and cell cycle regulation. Dysregulation of protein methylation has been closely linked to major diseases, including cancer, neurodegenerative disorders, and autoimmune diseases. Therefore, systematic and quantitative analysis of protein methylation patterns is critical for understanding cellular functional states and the molecular mechanisms of disease.
Leveraging advanced chromatography and mass spectrometry platforms, MtoZ Biolabs provides Methylation Proteomics Service for the identification and quantification of lysine and arginine methylation sites. This service supports researchers in elucidating the functional roles of protein methylation in biological processes such as signal transduction, chromatin remodeling, and transcriptional regulation. It is well-suited for basic research, disease mechanism studies, and drug mode-of-action analysis.
Analysis Workflow
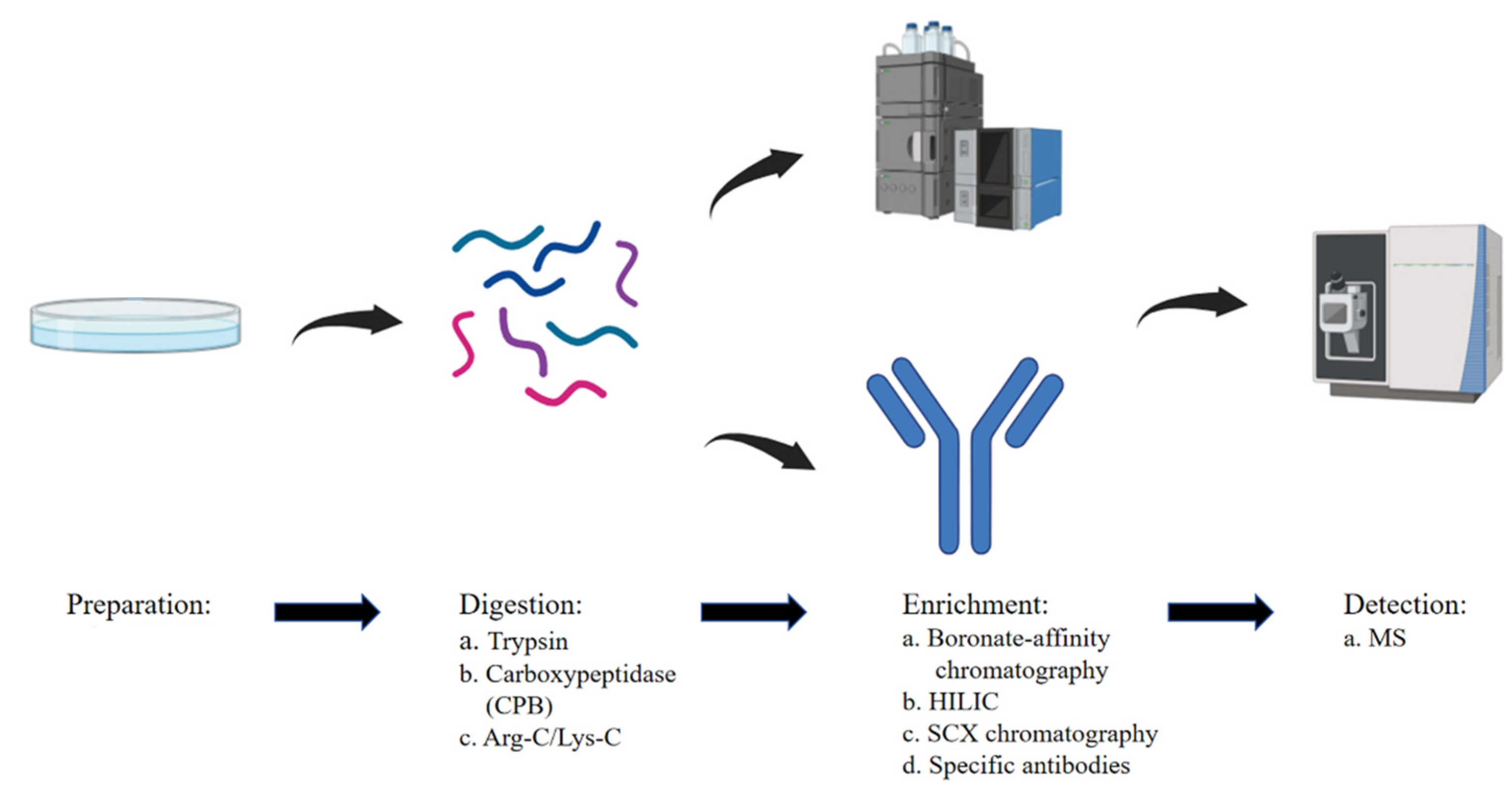
Wang X. et al. Cells. 2025.
The general workflow of Methylation Proteomics Service is as follows:
1. Sample Receipt and Quality Assessment
Samples such as cells, tissues, or body fluids are received and subjected to total protein extraction and quality control analysis.
2. Protein Digestion
Proteins are enzymatically digested into peptides using proteases such as trypsin.
3. Methylated Peptide Enrichment
Methylated peptides are enriched using immunoaffinity-based methods or chemical selective capture techniques.
4. LC-MS/MS Analysis
Enriched peptides are analyzed using high-resolution liquid chromatography–tandem mass spectrometry (LC-MS/MS).
5. Data Processing and Modification Identification
MS data is searched against protein databases to identify peptides and map methylation sites.
6. Bioinformatics Analysis
A comprehensive report is generated, including functional annotation, protein–protein interaction networks, and pathway analysis.
Service Advantages
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Methylation Proteomics Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all Methylation Proteomics data, providing clients with a comprehensive data report.
4. Supports Quantitative Comparative Experimental Design: Compatible with TMT, iTRAQ, and label-free strategies to meet differential expression analysis needs.
Sample Submission Suggestions
Sample Types: We accept a wide range of biological samples, including but not limited to frozen tissues, cell pellets, or protein extracts.
Storage Conditions: Samples should be stored at -80°C. Use dry ice or low-temperature cold chain during transportation to prevent degradation. Avoid repeated freeze-thaw cycles.
Preprocessing Requirements: If submitting protein extracts, please specify the buffer composition and indicate whether any denaturants or salts are present.
If you are unsure about sample preparation, MtoZ Biolabs offers preprocessing services and detailed guidance. Please contact our technical support team for assistance.
Applications
Example applications of Methylation Proteomics Service:
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Related Services
Post-Translational Modifications Proteomics Service
MS-Based Post-Translational Modification Analysis Service
How to order?







