Membrane Protein-Molecule Interaction Analysis Service
Membrane Protein-Molecule Interaction Analysis Service is an experimental service focused on investigating the binding characteristics between membrane proteins and small molecules, antibodies, proteins, or other bioactive molecules.
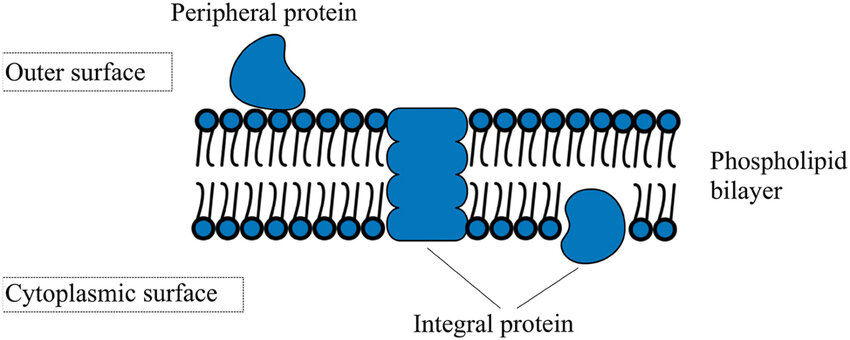
Ghazikhani H. Butler G. J Integr Bioinformat. 2023.
Membrane proteins play a central role in essential cellular activities, serving as key nodes in processes such as signal transduction, substance transport, and cell recognition. Notably, more than 60% of clinical drug targets are associated with membrane proteins. To investigate the binding behavior between membrane proteins and ligands—such as small molecules, antibodies, or proteins—real-time, label-free interaction analysis platforms have become indispensable tools in drug screening and mechanism elucidation.
Surface Plasmon Resonance (SPR) is a real-time, label-free optical sensing technology based on changes in the refractive index near a sensor surface. In this technique, membrane proteins or intact cells are immobilized on a metal-coated chip surface. When target molecules (e.g., small molecules, antibodies, or proteins) flow over the sensor surface, specific binding events alter the angle of reflected light, generating a sensorgram that captures the interaction in real time. By analyzing the association and dissociation phases, key kinetic parameters such as the association rate constant (kon), dissociation rate constant (koff), equilibrium dissociation constant (KD), and interaction specificity and stability can be obtained.
Leveraging an advanced SPR platform, MtoZ Biolabs offers Membrane Protein-Molecule Interaction Analysis Service to support the study of the binding characteristics of membrane proteins with various molecules under native or near-physiological conditions, comprehensively obtain key parameters such as affinity, kinetic constants (kon/koff) and binding specificity, and enable target validation, antibody screening, candidate drug screening and mechanism research.
Analysis Workflow
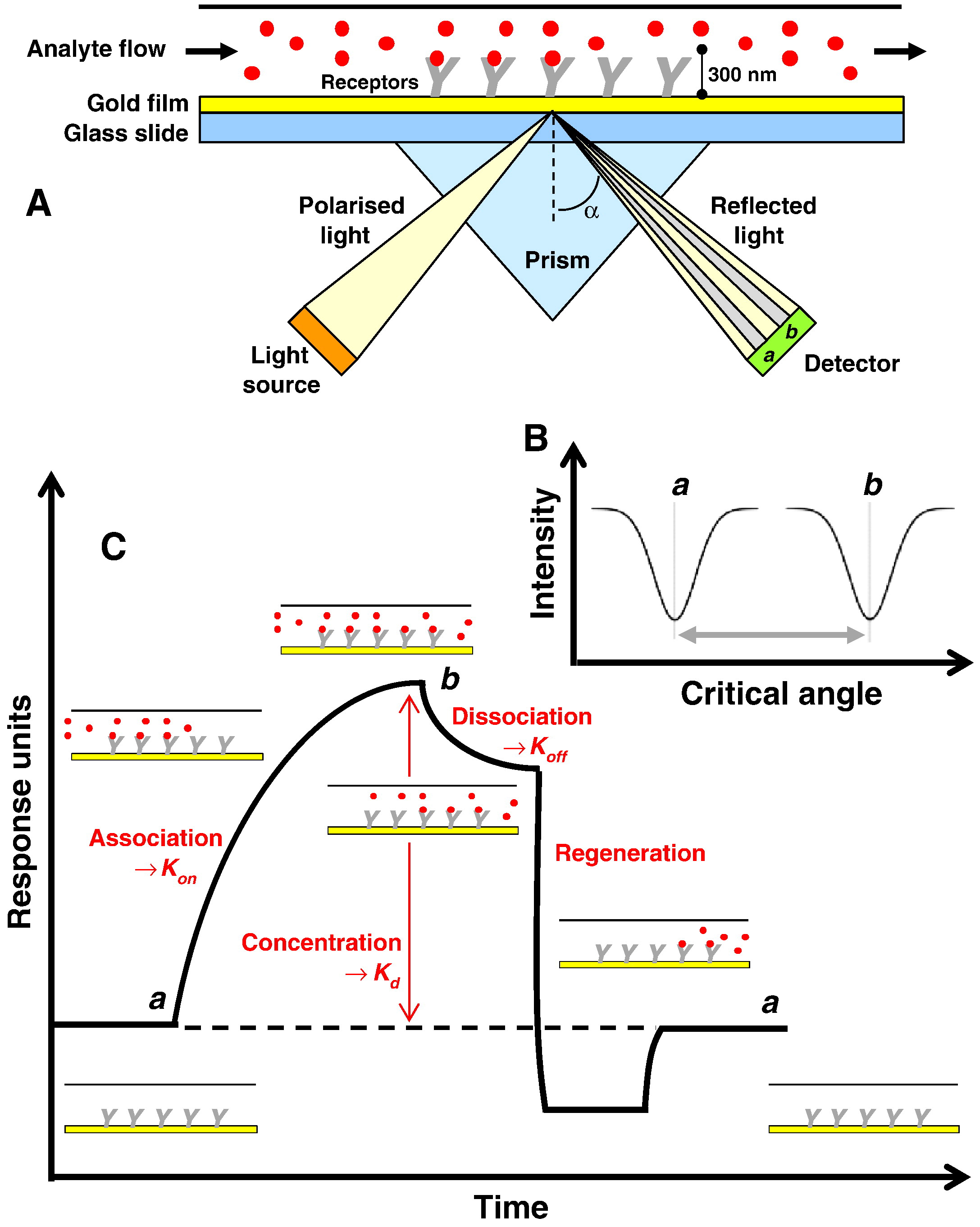
Patching SG. Biochim Biophys Acta. 2014.
The main workflow of Membrane Protein-Molecule Interaction Analysis Service is as follows:
1. Sample Immobilization
Membrane proteins are immobilized onto the surface of the sensor chip using an appropriate method.
2. Analyte Injection
The target molecules are injected over the chip surface via a microfluidic system with precise control of concentration and flow rate.
3. Real-Time Interaction Monitoring
The system continuously records the association and dissociation curves between the membrane protein and the analyte, reflecting the interaction process in real time.
4. Kinetic Parameter Calculation
Using multi-concentration fitting models, key kinetic parameters such as kon, koff, and KD are calculated.
5. Data Output and Analysis
A comprehensive report is provided, including raw sensorgrams, kinetic parameters, and analyses of binding stability and specificity.
Service Advantages
1. Real-Time, Label-Free Detection: Eliminates the need for fluorescent or radioactive labeling, preserving native binding behavior without structural interference.
2. Comprehensive Kinetic Profiling: Simultaneously captures kon, koff, and KD values, offering a complete kinetic perspective of molecular interactions.
3. Adaptability to Native Membrane Environments: Supports interaction studies in native membrane proteins or live cell systems.
4. High Sensitivity with Low Sample Requirements: Ideal for studies involving rare proteins, low-abundance membrane proteins, or scarce antibodies.
5. Excellent Reproducibility and High Standardization: Well-suited for large-scale drug or antibody screening applications.
Sample Submission Suggestions
Storage and Transportation: Purified membrane proteins should be stored at –80°C and shipped on dry ice. Test molecules (e.g., small molecules, antibodies) should be stored and transported at 4°C or –20°C, depending on their stability.
Special Samples: Please contact technical personnel before submission to confirm sample suitability and handling requirements.
Applications
Application examples of Membrane Protein-Molecule Interaction Analysis Service:
Validation of Candidate Drug–Target Binding
Analyze the binding kinetics between small-molecule drugs and membrane proteins for early-stage screening and optimization.
Antibody Affinity Measurement and Screening
Evaluate the binding specificity and stability of antibodies targeting transmembrane receptors to support antibody drug development.
Cell-Level Interaction Analysis
Enable experiments under immobilized live-cell conditions to study the binding behavior of cell surface proteins with drugs or antibodies.
Deliverables
1. Raw sensorgram plots
2. Kinetic parameter table including kon, koff, and KD
3. Binding specificity and stability analysis report
4. Documentation of experimental conditions and parameter settings
5. Technical summary and customized recommendations (if applicable)
Related Services
SPR Based Protein Interaction Analysis Service
How to order?







