SPR Based Protein Interaction Analysis Service
Developed in the 1990s, Surface plasmon resonance technology (SPR) offers an innovative methodology for detecting interactions between ligands and analytes on biosensor chips, grounded in SPR principles. Its applications span a wide range of fields, encompassing the study of various biomolecules such as peptides, proteins, oligonucleotides, and oligosaccharides, as well as the interactions among viruses, bacteria, cells, and small molecular compounds. This advanced and versatile technique plays a crucial role in deepening our understanding of biological processes and accelerating the development of novel therapeutics and diagnostics. Offering real-time, label-free analysis, SPR technology underscores its critical role in the meticulous investigation of biomolecular interactions, thus standing as a cornerstone in the biotechnological and biochemical research.
SPR involves the oscillation of longitudinal charge density at the interface between a metallic and a dielectric surface. This physical phenomenon is leveraged in SPR experiments, where one of the molecules under investigation is immobilized on the surface of a chip; this immobilized molecule is referred to as the ligand. Conversely, the other molecule, termed the analyte, flows over the chip's surface. Interactions between the analyte and the ligand lead to binding and dissociation events that result in changes in the mass at the chip surface, thereby altering the resonance angle. The sensorgram, recording these interaction-induced signal values in real-time, displays the binding kinetics and molecular affinity involved.
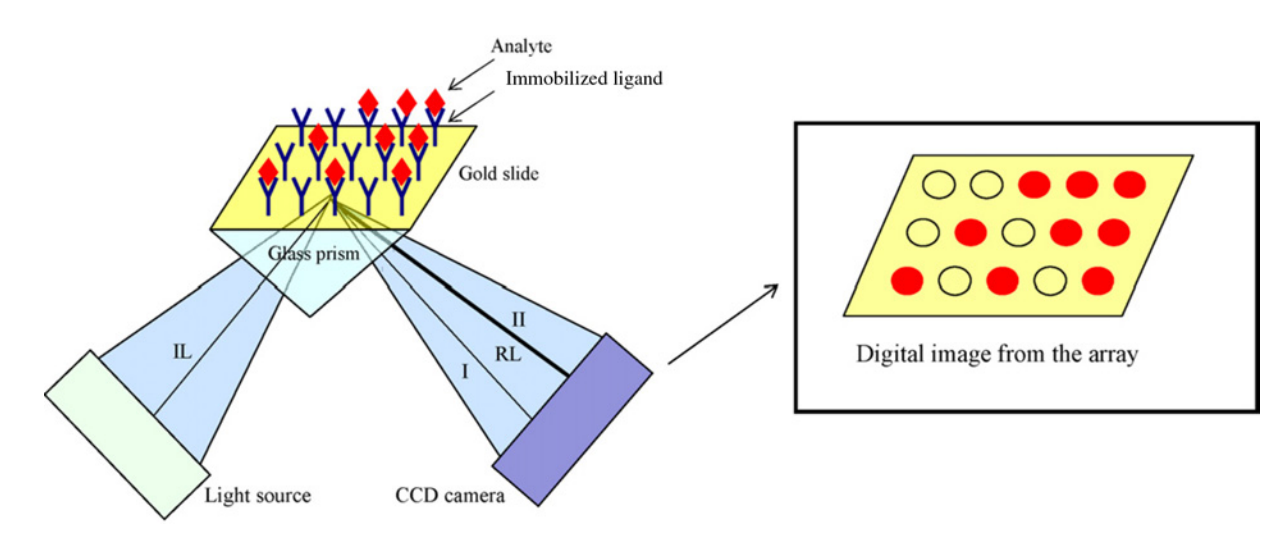
Figure 1. Schematic Representation of SPR
The principle of SPR experiments is based on the resonance between evanescent waves and surface plasmon waves on metal surfaces. This occurs when light transitions from a medium of higher optical density to one of lower optical density, under certain conditions, leading to total internal reflection. From the perspective of wave optics, when the incident light reaches the interface, it does not directly reflect. Instead, it penetrates the less dense medium to a depth of about one wavelength, then travels along the interface for about half a wavelength before returning to the denser medium, without a net change in the light's total energy. The wave that penetrates the less dense medium is known as an evanescent wave. Metals contain a sea of free electrons, which can be considered a type of plasma. The incident light excites longitudinal oscillations in this electron gas, generating charge density waves that propagate along the interface between the metal and the dielectric, forming surface plasmon waves. When resonance occurs between the surface plasmon waves and the evanescent waves, a significant reduction in the intensity of the reflected light is detected. With a fixed wavelength of incident light, the intensity of the reflected light is a function of the incident angle, with the lowest intensity corresponding to the resonance angle. SPR is highly sensitive to changes in the refractive index of the medium attached to the metal film surface. Changes in the properties of the surface medium or in the amount attached will result in a different resonance angle.
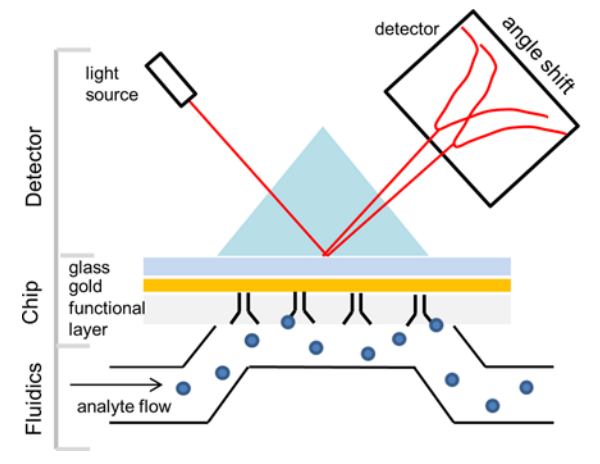
Figure 2. Schematic Diagram of SPR Principles [3]
The analysis cycle of SPR encompasses the following phases:
1. Baseline, which refers to the injection of the buffer solution.
2. Association, characterized by the continuous injection of the sample, where one.
observes the binding between the analyte and the ligand, generating a signal.
3. The introduction of the buffer solution leads to the dissociation of the analyte and ligand.
4. The injection of a regeneration solution completely elutes the analyte.
Subsequently, the buffer solution is injected again, returning to the baseline and entering the next analysis cycle. It is precise because of the feasibility of this cycle that SPR can be utilized for high-throughput screening (HTS) of pharmaceuticals.
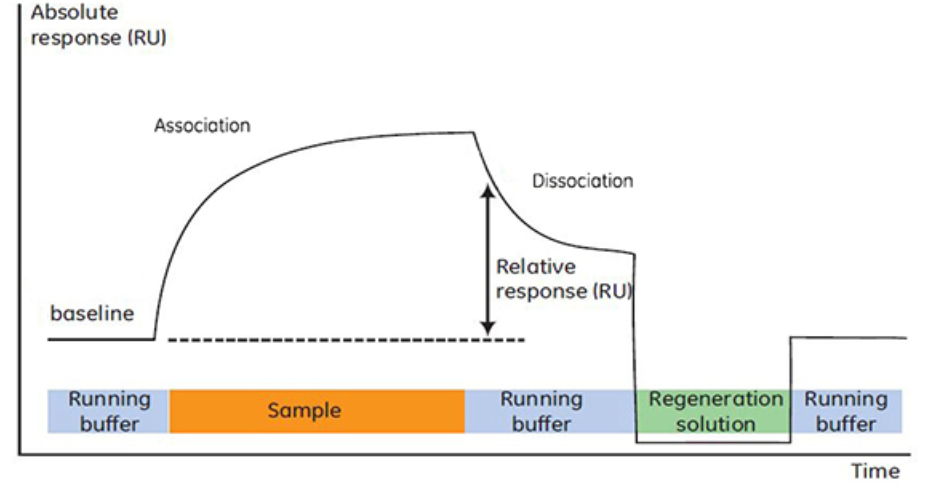
Figure 3. The Four Phases of SPR Experimental Analysis
SPR experiments can be categorized into two types: irreversible capture and reversible capture.
1. Irreversible Capture
Ligands are irreversibly bound to the chip surface. This approach utilizes amine, thiol, or aldehyde functional groups on proteins. The covalent coupling method utilizes exposed free carboxymethyl groups on the sensor chip's surface.
(1) Carboxy Methyl Dextran (CM5) for Amine Coupling
Suggested ligand concentration: 10-50 µg/mL (approximately 25 µg/mL). Maximum coupling density: 8,000-10,000 RU. It is recommended that the ligand's isoelectric point be above 4, as lower values are not conducive to amine coupling. This is the most commonly used chip type. It can be used to prepare capture chips (fixing antibodies or streptavidin).
(2) Streptavidin (SA) for Capturing Biotinylated Ligands
Amine coupling streptavidin to the CM5 chip can reduce nonspecific binding or achieve high coupling density. The Biotin CAPture kit can be selected for reversible capture. Recommended ligand concentration is approximately 2-5 µg/mL. Maximum capture capacity: 2,000 RU.
2. Reversible Capture
Ligands are removable from the chip surface each cycle. Currently, three capture methods are commonly used: capture of biotinylated molecules on streptavidin sensor chips, capture of His-tagged proteins by nickel-nitrilotriacetic acid (Ni-NTA) groups, and capture by immobilized specific antibodies.
(1) Ni-NTA for Capturing His-tagged Proteins
Suggested ligand concentration is approximately 10 µg/mL. Maximum capture capacity: 1,000-3,000 RU, with 8 His or 10 His-tagged proteins providing a larger coupling amount (up to 5,000 RU).
(2) Biotin CAPture Kits for Capturing Biotinylated Ligands
Suggested ligand concentration is approximately 2-5 µg/mL. The chip is pre-immobilized with specific single-stranded DNA, and the Biotin CAPture kit provides biotinylated complementary DNA.
(3) Antibody Capture Kits
Anti-human, Anti-mouse, Anti-GST, and Anti-His capture kits offer the corresponding antibodies and reagents, which can be immobilized onto the CM5 chip through amine coupling for reversible antibody capture. The maximum immobilization quantity depends on the antibody concentration.
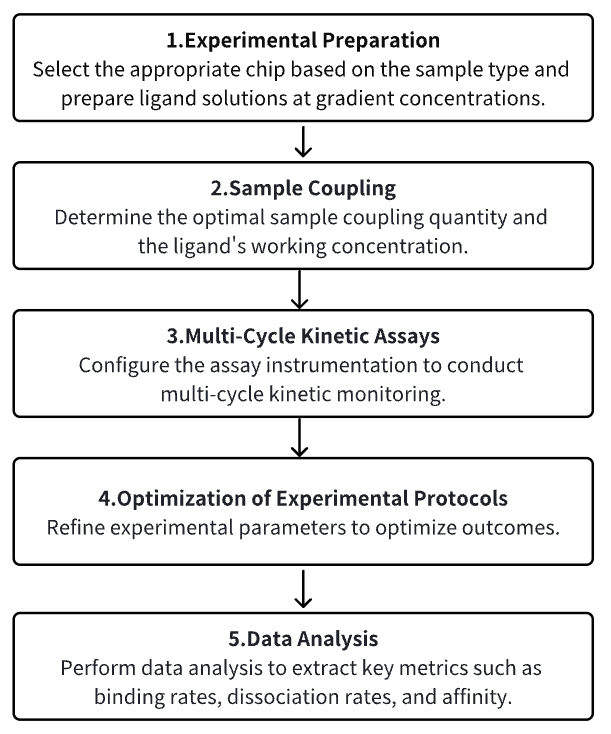
Analysis Workflow
1. Experimental Preparation
2. Sample Coupling
3. Multi-Cycle Kinetic Assays
4. Optimization of Experimental Protocols
5. Data Analysis
Service Advantages
1. Enables detection across a broad spectrum of samples, encompassing interactions between proteins, peptides, and small molecules.
2. Offers a diverse array of services, encompassing biomolecular interaction analysis, antibody screening, and nucleic acid aptamer selection, among others.
3. Delivers highly reliable detection outcomes, complemented by comprehensive bioinformatics analysis.
Sample Results
1. SPR Detection of the Interaction between Silver Nanoparticles and Proteins [4]
The application of silver nanoparticles in biological systems is increasingly prevalent, for instance, as potential candidates for antimicrobial agents and controlled drug release systems. Consequently, researching these interactions is of significant importance. While BSA has been extensively used as a model protein to study interactions with silver nanoparticles, studies using other proteins are considerably limited. The interaction of silver nanoparticles with light leads to the collective oscillation of conduction electrons, resulting in SPR. By investigating the protein concentration dependency of SPR band spectra for numerous proteins, it was found that for all proteins, as the concentration increases, the SPR band intensity initially decreases, reaches a minimum value, and then increases again, creating a characteristic "dip and rise" pattern. The nadir of this pattern seems to be related to the isoelectric point of the protein. Detailed dynamic light Scattering and transmission electron microscopy studies indicate that the consistency of SPR spectra depends on the average particle size of silver nanoparticles and their associated states with changes in protein concentration.
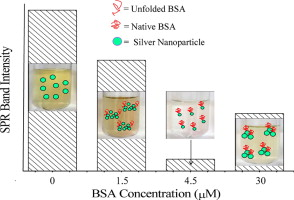
Figure 4. Interaction between Silver Nanoparticles and Proteins
2. SPR Detection of the Interaction between Schistosome Esophageal MEG Proteins and Human S100 Proteins Involved in Inflammatory Responses [5]
The micro-exon gene-14 (MEG-14) displays significant structural features that allow for antigenic variation production in schistosomes. The soluble portion of MEG-14 proteins exhibits characteristics of intrinsically disordered proteins and is expressed only in the parasite's esophageal glands. These characteristics suggest the possibility of interaction with host proteins present in ingested blood plasma and cells. This study employed pull-down and SPR experiments to verify the interaction between Schistosoma mansoni MEG-14 (sMEG-14) and human S100A9 proteins.
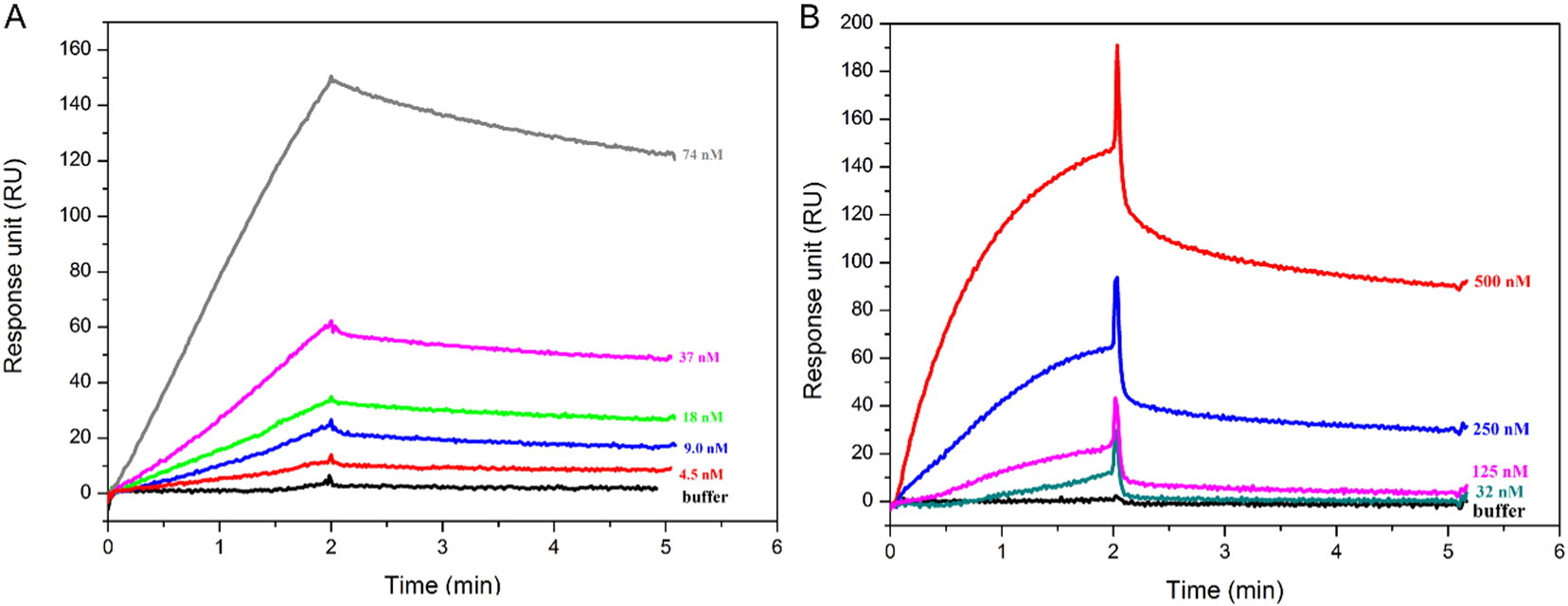
Figure 5. SPR Analysis of the Binding between S100A9 or S100A8/A9 and sMEG-14
3. Evaluating the Interaction between Pesticides and Cell Membrane Models through SPR Spectroscopy [6]
SPR spectroscopy is utilized to characterize the interaction between pesticides and cell membrane models. Liposomes are immobilized on the SPR sensor chip (L1) surface, with the lipid bilayer formed on the sensor chip considered as a model for cell membranes. A solution containing pesticides is flowed over the sensor chip, yielding a SPR sensor graph that reflects the interaction between the pesticides and the lipid bilayer. The patterns and strengths of the interaction between the pesticides and the cell membrane model can be visualized and quantified through SPR. Triflumizole, hexaconazole, and pentachlorophenol exhibit strong interactions with the lipid bilayer.
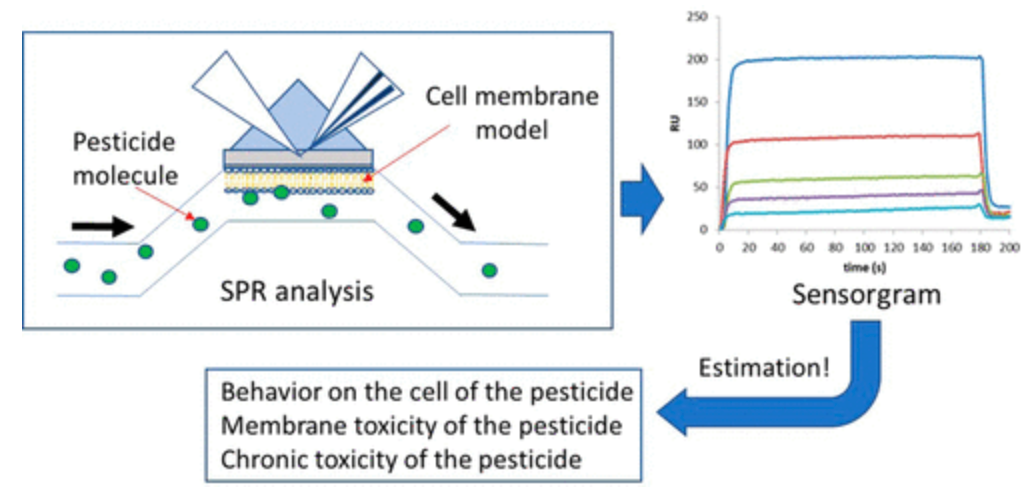
Figure 6. SPR Evaluation of the Interaction between Pesticides and Cell Membrane Models
4. Multiplexed Analysis of Macromolecular Interactions via SPR Imaging: Application to p53 and DNA Interactions [7]
SPR is an optical technique where the binding of analytes to a surface alters the refractive index at the surface/solution interface. Molecular interactions can be analyzed in real-time without the need for labeling steps. Currently, the limitation of SPR imaging is the few reactions that can be analyzed simultaneously. By employing novel grafting techniques and a new imaging system, we have enhanced the throughput of SPR imaging. The interaction between p53 and DNA was chosen as a paradigm to validate this assay. Using a labeled DNA approach, we targeted multiple DNA sequences on a single chip simultaneously. The interaction between p53 and these DNA sequences was monitored via SPR imaging. Both qualitative and quantitative analyses provided results analogous to those obtained using traditional techniques.
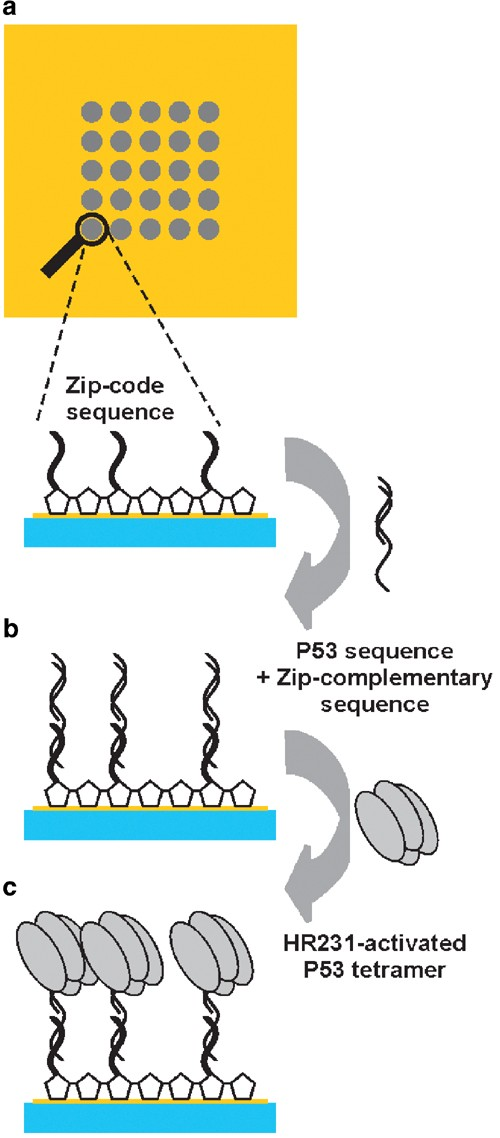
Figure 7. SPR Experiments for Detecting p53 DNA Binding Activity
Sample Submission Requirements
1. Please provide samples that are as purified as possible.
2. Endeavor to minimize contamination with impurities.
Services at MtoZ Biolabs
1. Complete Experimental Procedures
2. Relevant Instrument Parameters
3. Original SPR Data
4. Detailed Report on Sample Interactions (Including Kinetic Parameters (Association and Dissociation Rates), Affinity (Equilibrium Dissociation Constant, Kd), etc.)
Applications
1. SPR for Detection of Food Allergens in Food Matrices [8]
Food allergies, immune-mediated adverse systemic reactions, are now considered a global health issue, increasingly common in the realm of food safety. Food allergies can stimulate the immune system and induce an IgE-mediated hypersensitivity reaction, releasing allergic inflammatory mediators and triggering severe acute hypersensitivity reactions, such as rashes, hives, diarrhea, abdominal pain, and even anaphylactic shock. SPR biosensors, due to their extreme sensitivity to reactants (including food allergens, toxic proteins, labeled proteins, antibodies, and drug proteins), have been utilized in the detection of food allergens.
2. SPR in Target-Based Drug Development
Taking COVID-19 as an example, the majority of efforts to find or develop potential treatments for COVID-19 are based on the role of the ACE2 protein in allowing the virus to enter host cells, testing new and existing compounds by blocking the interaction between host cells and ACE2. Studies have identified potential small molecules that inhibit ACE2 binding and have evaluated the binding affinity and kinetics of these potential therapeutic small molecules to ACE2 protein using SPR.[9]
3. The Role of SPR in Antibody-Based Drug Discovery
SPR for affinity/kinetic measurements of candidate drugs has always been a primary application in characterizing candidate drugs. Recently, with the growth of monoclonal antibodies (mAbs) as a drug category and the availability of higher throughput biosensors, the role of label-free biosensors has evolved to include epitope mapping and mAb positioning in the early drug discovery process through epitope characterization.
4. Clinical Biomarker Detection [10]
SPR can perform target analysis in molecular diagnostics in complex samples (such as serum, saliva, blood, and urine), with analytes including hormones (steroids and peptides), protein clinical biomarkers, antibodies involved in immune diseases, nucleic acids for genetic diseases, clinical biomarkers (i.e., miRNA), bacterial cells, and viruses for pathogen detection.
FAQ
Q1: How should the chip for an SPR experiment be selected?
The selection should be based on the type of sample. For instance, within the BIAcore chip series, CM5 is suitable for protein samples. CM7 is tailored for small molecular samples with a coupling capacity three times that of CM5. It is a requisite choice when the molecular weight ratio of large molecules to small molecules exceeds 500. SA/NA chips are designed for biotin-labeled samples (employing the irreversible binding principle of streptavidin and biotin; for reversible requirements, opt for the Biotin CAPture Kit). NTA chips are appropriate for samples with HIS tags. Protein A/G chips are ready-to-use and suitable for antibodies.
Q2: What kinds of interactions can SPR study?
In principle, SPR can be applied to the interaction of any type of molecules, ranging from organic compounds to proteins, nucleic acids, glycoproteins, and even viruses and whole cells. Since the response measures changes in mass concentration, the response per mole of interacting substances is directly proportional to the molecular weight (the smaller the molecule, the lower the molar response).
References
[1] Chan HCS, Shan H, Dahoun T, Vogel H, Yuan S. Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol Sci. 2019 Aug;40(8):592-604. doi: 10.1016/j.tips.2019.06.004. Epub 2019 Jul 15. Erratum in: Trends Pharmacol Sci. 2019 Oct;40(10):801. PMID: 31320117.
[2] Pinzi L, Rastelli G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int J Mol Sci. 2019 Sep 4;20(18):4331. doi: 10.3390/ijms20184331. PMID: 31487867; PMCID: PMC6769923.
[3] Damborský, Pavel & Švitel, Juraj & Katrlík, Jaroslav. (2016). Optical biosensors. Essays In Biochemistry. 60. 91-100. 10.1042/EBC20150010.
[4] Banerjee V, Das KP. Interaction of silver nanoparticles with proteins: a characteristic protein concentration dependent profile of SPR signal. Colloids Surf B Biointerfaces. 2013 Nov 1;111:71-9. doi: 10.1016/j.colsurfb.2013.04.052. Epub 2013 May 25. PMID: 23792543.
[5] Orcia D, Zeraik AE, Lopes JLS, Macedo JNA, Santos CRD, Oliveira KC, Anderson L, Wallace BA, Verjovski-Almeida S, Araujo APU, DeMarco R. Interaction of an esophageal MEG protein from schistosomes with a human S100 protein involved in inflammatory response. Biochim Biophys Acta Gen Subj. 2017 Jan;1861(1 Pt A):3490-3497. doi: 10.1016/j.bbagen.2016.09.015. Epub 2016 Sep 15. PMID: 27639541.
[6] Moriwaki H, Yamada K, Nakanishi H. Evaluation of the Interaction between Pesticides and a Cell Membrane Model by Surface Plasmon Resonance Spectroscopy Analysis. J Agric Food Chem. 2017 Jul 5;65(26):5390-5396. doi: 10.1021/acs.jafc.7b01895. Epub 2017 Jun 20. PMID: 28602084.
[7] Maillart E, Brengel-Pesce K, Capela D, Roget A, Livache T, Canva M, Levy Y, Soussi T. Versatile analysis of multiple macromolecular interactions by SPR imaging: application to p53 and DNA interaction. Oncogene. 2004 Jul 15;23(32):5543-50. doi: 10.1038/sj.onc.1207639. PMID: 15184889.
[8] Zhou J, Qi Q, Wang C, Qian Y, Liu G, Wang Y, Fu L. Surface plasmon resonance (SPR) biosensors for food allergen detection in food matrices. Biosens Bioelectron. 2019 Oct 1;142:111449. doi: 10.1016/j.bios.2019.111449. Epub 2019 Jun 21. PMID: 31279816.
[9] Pan B, Fang S, Zhang J, Pan Y, Liu H, Wang Y, Li M, Liu L. Chinese herbal compounds against SARS-CoV-2: Puerarin and quercetin impair the binding of viral S-protein to ACE2 receptor. Comput Struct Biotechnol J. 2020;18:3518-3527. doi: 10.1016/j.csbj.2020.11.010. Epub 2020 Nov 11. PMID: 33200026; PMCID: PMC7657012.
[10] Mariani S, Minunni M. Surface plasmon resonance applications in clinical analysis. Anal Bioanal Chem. 2014 Apr;406(9-10):2303-23. doi: 10.1007/s00216-014-7647-5. Epub 2014 Feb 25. PMID: 24566759; PMCID: PMC7080119.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Active Substances Screening Service
How to order?







