iTRAQ-Based Peptidomics Analysis Service
iTRAQ (isobaric tags for relative and absolute quantification) is an isobaric labeling technique that enables chemical labeling of different samples after peptide digestion, thereby allowing for simultaneous detection and relative quantification of multiple samples. This service labels each sample with a distinct isobaric tag, resulting in identical mass in the MS1 stage while releasing signature reporter ions in the MS/MS stage for quantitative analysis. This analytical approach is a high-throughput and quantitatively precise strategy for multi-sample comparison, particularly suitable for investigating expression differences of endogenous peptides with low to medium molecular weight.
The iTRAQ-based peptidomics analysis service is widely applied to differential expression studies of various biological samples, especially for high-throughput quantitative analysis of low- to medium-molecular-weight peptides in tissue fluids, body fluids, and digestive fluids. It is commonly used to explore disease-associated peptides, identify potential biomarkers, investigate cell signaling pathways, study pharmacological mechanisms, and analyze functional components of food-derived peptides, making it an essential quantitative tool in peptidomics research.
Services at MtoZ Biolabs
Based on a high-resolution mass spectrometry platform, MtoZ Biolabs offers an iTRAQ-based peptidomics analysis service that enables simultaneous relative quantification of peptides across multiple samples. This service integrates iTRAQ isobaric labeling technology with advanced LC-MS/MS systems to perform high-throughput and quantitatively accurate expression profiling of endogenous peptides in complex samples such as plasma, tissues, cells, and body fluids. It provides clients with high-quality peptidomic data to support in-depth investigation of peptide expression dynamics under physiological and pathological conditions.
Analysis Workflow
1. Sample Preparation and Peptide Extraction
Endogenous peptides are extracted from plasma, tissues, or cells by removing high-molecular-weight proteins while retaining low-molecular-weight peptides.
2. iTRAQ Labeling
Peptides from different sample groups are labeled with iTRAQ reagents, enabling multiplexed parallel analysis.
3. High-Resolution Mass Spectrometry Detection
Accurate peptide spectra and quantitative data are acquired using LC-MS/MS platforms such as Orbitrap or TripleTOF.
4. Data Processing and Quantitative Analysis
Peptide identification, ratio calculation, and statistical analysis are performed using specialized software to generate lists of differentially expressed peptides and related functional annotations.
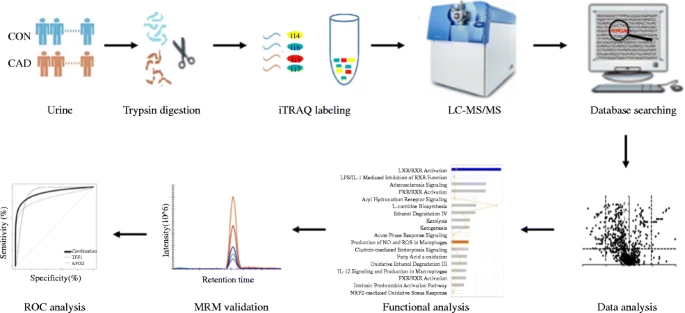
Sun, H D. et al. Analytical and Bioanalytical Chemistry, 2019.
Figure 1. The Workflow of the iTRAQ Analysis.
Sample Submission Suggestions
1. Sample Types
Various biological samples are supported, including plasma, serum, tissue homogenates, and cell lysates from human or animal sources. Samples should be free of lipids and impurities, with precautions taken to prevent protein degradation.
2. Sample Storage and Transportation
Samples should be stored at low temperatures (−80 °C) and transported on dry ice under cold chain conditions. Detailed guidelines for sample handling and packaging are also available to assist clients in standardizing procedures.
Service Advantages
1. Professional Platform Ensures Data Quality
Powered by MtoZ Biolabs’ high-resolution mass spectrometry platform and standardized workflows, this service guarantees reproducibility and accuracy, supporting large-scale peptidomics research projects.
2. High Throughput and Quantitative Accuracy
Utilizing isobaric labeling and tandem mass spectrometry, this approach offers full-spectrum coverage and precise quantification, suitable for detecting subtle expression changes in complex samples.
3. Simultaneous Multi-Sample Analysis
iTRAQ labeling enables the parallel analysis of multiple samples, allowing cross-group and time-point comparisons that enhance research efficiency and statistical reliability.
4. Customized Service Solutions
Tailored experimental designs are available based on client objectives, including labeling strategies, sample grouping, and data analysis configurations, helping researchers achieve targeted scientific outcomes.
Applications
1. Analysis of Biological Regulatory Mechanisms
By profiling regulatory peptides associated with key cellular processes such as proliferation, differentiation, and apoptosis, this service facilitates the exploration of their functional roles and action pathways.
2. Signal Transduction Pathway Investigation
iTRAQ-based peptidomics analysis service enables dynamic expression profiling of signaling or regulatory peptides under varying conditions, supporting the understanding of cellular communication and regulatory networks.
3. Biomarker Discovery and Validation
Stable and reliable peptide candidates can be identified in biofluids such as plasma, cerebrospinal fluid, and urine, aiding in early disease screening or therapeutic efficacy evaluation.
4. Nutrition and Metabolic Regulation Studies
iTRAQ-based peptidomics analysis service supports the assessment of endogenous peptide responses to dietary intake or metabolic imbalance, informing the development of nutritional intervention strategies.
FAQ
Q1: Is iTRAQ Labeling Applicable to All Types of Peptides?
A1: iTRAQ tags label the N-terminus and lysine residues of peptides, making them suitable for most peptides. However, labeling efficiency can be affected by amine-containing contaminants in the sample or suboptimal proteolysis. Therefore, it is recommended to provide well-digested samples with minimal background interference.
Q2: Does iTRAQ Labeling Interfere with Subsequent Mass Spectrometry Detection?
A2: iTRAQ tags have a small molecular weight and exert minimal impact on mass spectrometry separation and detection. With optimized sample preparation and detection workflows, high sensitivity and quantification accuracy can be reliably achieved.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
How to order?







