HLA Peptidomics Analysis Service
HLA peptidomics analysis is a research method that uses mass spectrometry to systematically identify and quantify endogenous peptides bound to human leukocyte antigen (HLA) molecules, aiming to reveal the mechanisms of antigen presentation and their roles in immune recognition. This technique enriches HLA-bound peptides from HLA class I or class II molecules through immunoaffinity purification and performs sequence identification and abundance analysis using high-resolution LC-MS/MS platforms to obtain peptide characteristics and binding specificity information.
HLA peptidomics analysis service has broad application value in fields such as tumor immunology, vaccine development, and immune pathway research. By comprehensively analyzing the antigen peptide repertoire presented on HLA molecules, researchers can identify potential targets related to immune responses, supporting the discovery of novel T cell epitopes, tumor neoantigen recognition, and the development of precise immunological strategies.
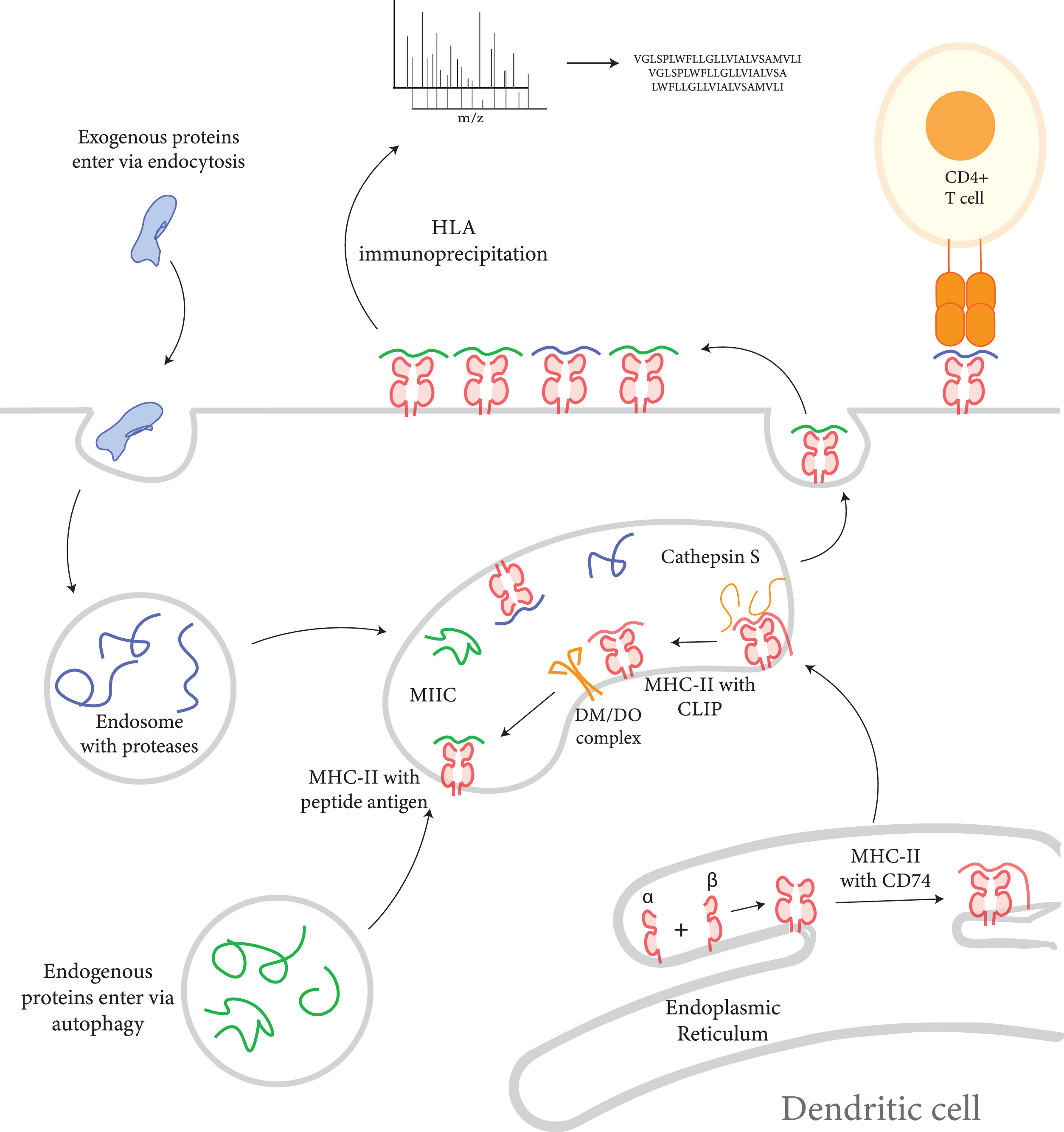
Taylor, H B. et al. Molecular & Cellular Proteomics, 2021.
Figure 1. Profiling the HLA-II Processing and Presentation Pathway by LC-MS/MS.
Services at MtoZ Biolabs
Based on high-resolution mass spectrometry platforms, MtoZ Biolabs offers the HLA peptidomics analysis service focusing on the systematic analysis of endogenous peptides presented by HLA molecules. This service uses immunoaffinity enrichment to extract peptides, followed by LC-MS/MS for sequence identification and relative abundance analysis. It provides information on HLA-bound peptide sequences, modification status, abundance distribution, and potential immune-related functions, delivering critical data support for further research. The services provided include, but are not limited to, the following:
1. HLA Peptide Profiling
Comprehensively identifies and quantifies endogenous peptides presented by HLA molecules, outlining sample-specific antigenic peptide repertoires.
2. HLA Peptide Binding Assay
Evaluates the interactions between HLA molecules and specific peptides.
3. HLA Typing
Determines HLA class I and II types in the sample, supporting antibody selection for peptide enrichment and providing essential data for peptide binding prediction, T cell epitope identification, and immunological analysis.
4. HLA Peptide Elution
Utilizes immunoaffinity purification to elute HLA-bound peptides from cells or tissues, ensuring specificity for downstream mass spectrometry analysis.
5. HLA Epitope Analysis
Predicts and analyzes potential T cell epitopes, assessing their immunogenicity and presentation potential under specific HLA contexts.
6. LC-MS/MS Analysis of HLA Peptidome
Applies high-resolution mass spectrometry to accurately identify and quantify HLA-bound peptides, delivering high-quality sequence and abundance data.
Analysis Method
1. HLA Peptide Enrichment
Uses immunoaffinity purification with specific antibodies to selectively elute HLA-bound peptides from cells or tissues.
2. Mass Spectrometry Analysis (LC-MS/MS)
Performs high-sensitivity detection of eluted peptides using liquid chromatography–tandem mass spectrometry, capturing accurate mass-to-charge ratio data.
3. Peptide Sequencing
Interprets HLA peptide sequences from mass spectrometry data through database searching or de novo algorithms, identifying antigen origins and HLA binding features.
4. Data Annotation and Epitope Prediction
Integrates HLA typing and mass spectrometry results to identify peptides, predict HLA binding, annotate T cell epitopes, assess immunogenicity, and generate comprehensive immune-related data reports.
Service Advantages
1. High-Resolution Mass Spectrometry Platform
Utilizes advanced mass spectrometers such as Orbitrap, offering excellent sensitivity and resolution for accurate detection of low-abundance HLA-bound peptides.
2. High-Specificity Peptide Enrichment
Employs anti-HLA monoclonal antibodies for immunoaffinity purification, ensuring that eluted peptides originate from authentic HLA-peptide complexes, enhancing data reliability.
3. One-Time-Charge
Our pricing is transparent, no hidden fees or additional costs.
4. Comprehensive Workflow Coverage
Delivers end-to-end services from HLA typing, peptide enrichment, LC-MS/MS analysis to epitope annotation and functional interpretation, supporting in-depth mechanistic research.
Applications
1. Tumor Neoantigen Identification
HLA peptidomics analysis service can be used to analyze peptides presented by HLA molecules in tumor tissues, uncovering potential neoantigens to support immune target discovery.
2. Vaccine Epitope Screening
By identifying antigenic peptides with high HLA-binding affinity, this service facilitates the design of more immunogenic epitopes for viral, bacterial, or tumor vaccines.
3. T Cell Response Mechanism Study
HLA peptidomics analysis service enables identification of HLA-restricted peptides recognizable by T cells, helping to explore immune activation pathways and antigen presentation features.
4. HLA Binding Specificity Analysis
Investigates the peptide-binding preferences of different HLA alleles, supporting studies on HLA-peptide interaction mechanisms and structure–function relationships.
FAQ
Q1: How Is HLA Peptidomics Analysis Different from Conventional Proteomics?
A1: HLA peptidomics focuses on identifying endogenous peptides bound to HLA molecules and is primarily concerned with antigen presentation. In contrast, conventional proteomics emphasizes the expression levels and functional annotation of whole proteins or peptides. The two approaches differ significantly in both research objectives and analytical workflows.
Q2: Can Modified Peptides Be Identified?
A2: Yes. We support the identification and annotation of post-translationally modified HLA-bound peptides, such as phosphorylation, acetylation, and methylation. However, this should be specified in advance to optimize detection workflows and data analysis settings.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Data Analysis, Preprocessing, and Estimation
4. Bioinformatics Analysis
5. Raw Data Files
How to order?







