Protein Corona Characterization Service
Due to their large surface area-to-volume ratio, nanoparticles (NPs) rapidly adsorb proteins on their surface upon contact with biological fluids, forming a "protein corona (PC)." The binding between NPs and proteins is primarily mediated by intermolecular forces such as hydrogen bonding, hydrophobic interactions, electrostatic interactions, and van der Waals forces. During this process, proteins with strong binding affinity can quickly form a so-called "hard PC" on the surface of NPs, surrounded by a "soft PC" with weaker interactions. Hard PCs are more diverse and stable, with longer residence times. Soft PCs have weaker binding to NPs and are easily replaced by free proteins in the culture medium, requiring hours or even days to reach equilibrium, with higher exchange rates and shorter residence times. Over time, proteins with stronger affinity gradually replace those with weaker affinity, a phenomenon known as the Vroman effect.
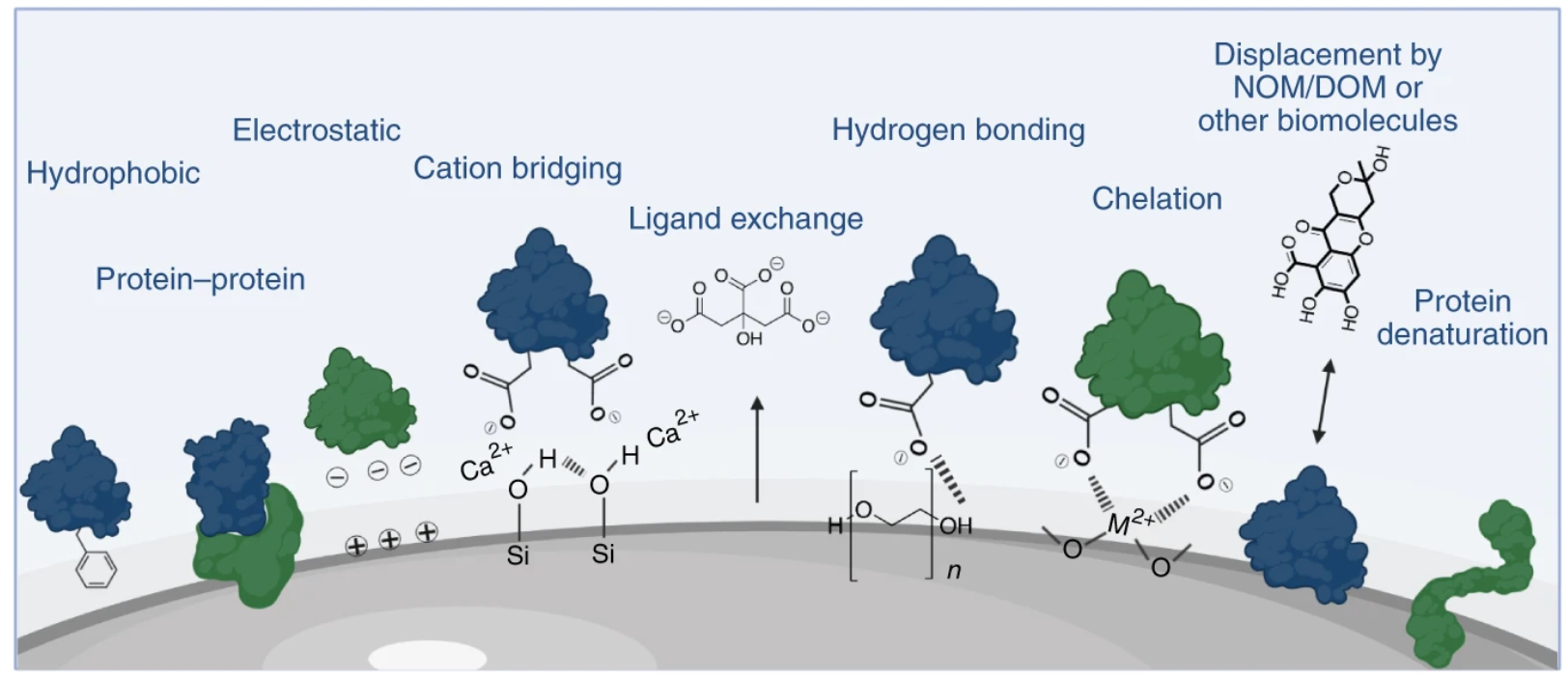
Figure 1. Surface Interactions Mediating PC Formation[3]
In addition to protein characteristics, the formation of PCs is influenced by the physicochemical properties of NPs (size, shape, surface modifications) and environmental conditions (temperature, pH, incubation time). (1) Chemical composition: Among the factors influencing PC formation, the chemical composition of NPs is the most important factor. For example, studies have shown that the serum PCs formed around nano-silver particles differ from those around nano-gold particles with the same surface ligand modification, with only 36.9% of protein types being the same. (2) Size: The size of NPs is closely related to surface curvature and is one of the most important factors influencing PC formation. On one hand, particle size directly affects changes in the corresponding surface area; on the other hand, proteins undergo different conformational changes on the surfaces of NPs with different curvatures, ultimately affecting the quantity, stability, and equilibrium time of adsorbed proteins. (3) Shape: The formation of PCs also depends on the shape of NPs, which can alter the structure, types, and quantities of proteins bound to their surfaces. (4) Surface modifications: Surface modifications of NPs can alter their surface charge and hydrophobicity/hydrophilicity. Surface charge determines the quantity of proteins adsorbed onto NPs; compared to positively charged NPs, neutrally charged NPs generally bind fewer proteins. In general, plasma proteins are negatively charged under physiological conditions with a pH of 7.4; therefore, positively charged NPs in blood tend to adsorb more proteins than negatively charged ones. In addition to surface charge, the hydrophobicity or hydrophilicity of NPs is also a key factor influencing PC composition. Compared to hydrophilic surfaces, hydrophobic surfaces exhibit greater affinity for proteins and a stronger tendency to denature adsorbed proteins. This may be due to hydrophobic interactions between hydrophobic molecules on the NP surface and proteins, promoting protein unfolding.
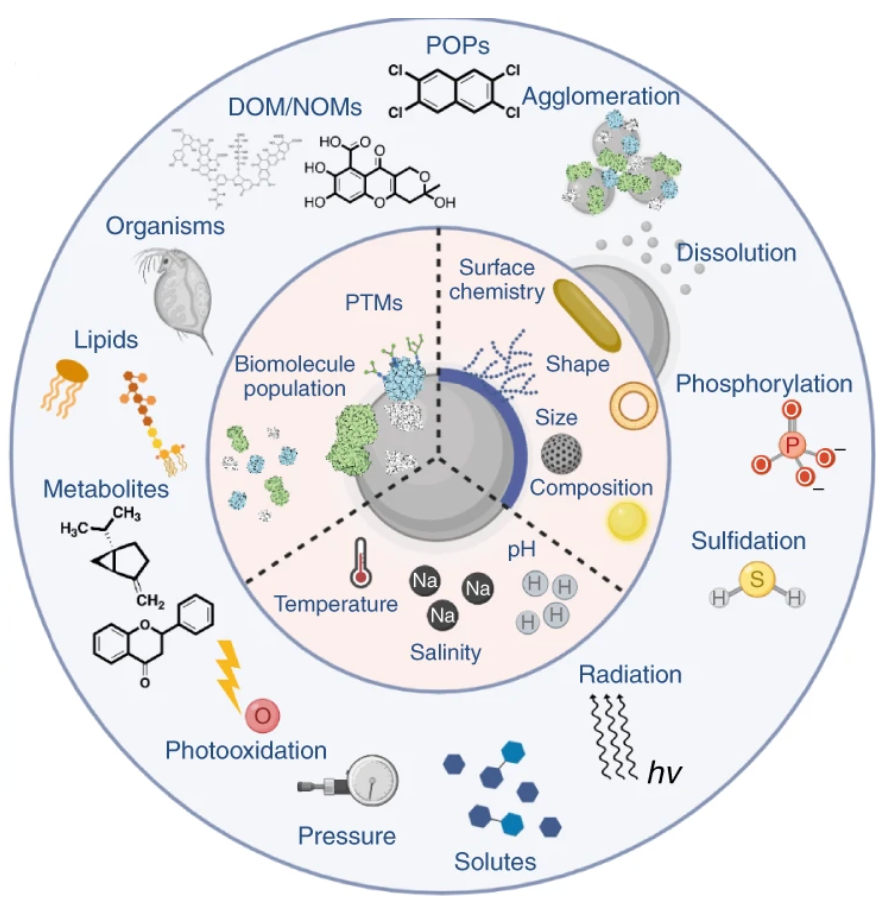
Figure 2. Factors Influencing PC Formation[3]
Environmental conditions also affect PC formation. (1) Composition of the culture medium: Culture media contain thousands of proteins that compete for adsorption onto the surface of NPs. PCs typically contain 10–50 proteins with high affinity for NPs. Differences in small molecule components such as sugars, lipids, or salts in the culture medium result in different PCs on the surface of NPs. (2) Incubation time: Although PCs rapidly form on the surface of NPs upon contact with biological fluids, their composition dynamically changes over time until a stable inner hard PC is formed. (3) Temperature: Fluctuations in environmental temperature also have different effects on PC formation. On one hand, temperature affects protein diffusion and affinity for NPs. Investigating these subtle temperature changes is crucial for understanding their impact on the adsorption of proteins onto NP surfaces. (4) Environmental pH: Environmental pH not only controls the protonation state of NP surfaces, thereby affecting their aggregation tendency, but also influences the binding affinity between NPs and proteins, further altering protein adsorption patterns. During biological absorption, nanomaterials experience various liquid environments with different pH values, such as exposure media (pH 6.9–7.4), blood (pH 7.35–7.45), intracellular fluid (pH 6.8–7), and lysosomes (pH 4.5–5). As the pH of the exposure solution gradually approaches the isoelectric point of proteins, protein stability in the solution decreases, leading to aggregation and adsorption onto available NP surfaces.
1. Analysis Techniques for PC
PCs are typically formed by weak interactions, and due to their complex and time-dependent kinetic behavior, no single technique can fully characterize all properties of protein-NP interactions, including particle size distribution, density, composition, molecular weight of molecules, etc. Many techniques can be employed for in-depth analysis of PCs, making it crucial to select appropriate techniques.
(1) Size Analysis
Ultracentrifugation (UC), Differential Centrifugal Sedimentation (DCS), Asymmetrical Flow Field-Flow Fractionation (AF4), and Light Scattering-Based Methods (DLS) are commonly used techniques for analyzing changes in NP size during PC formation. Combining these methods with DLS can provide more accurate particle size measurement results. Data obtained from AF4, DLS, and DCS measurements can be used to determine the apparent density of PCs of different NP/protein masses. However, this technique cannot determine the dry thickness of the protein layer, and even with different morphologies and shapes, results need to be interpreted based on the hydrodynamic size of spherical NPs.
The monodispersity and size of inorganic colloids can be characterized using Small-Angle X-ray Scattering (SAXS) techniques, while Transmission Electron Microscopy (TEM) can be used for solid-state characterization. By comparing NPs before and after corona formation, the thickness and size of the protein layer can be evaluated. However, due to the low contrast of organic materials, SAXS and TEM are not effective for characterizing the organic layer on the surface of nanoscale inorganic NPs. Small-Angle Neutron Scattering (SANS) is a powerful tool for selectively characterizing the organic and inorganic components of mixed NPs.
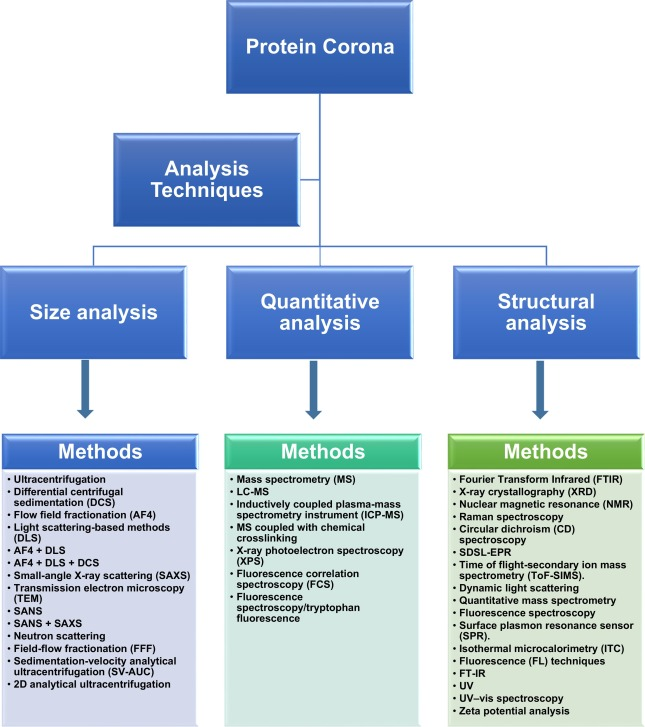
Figure 3. Experimental Techniques for PC Analysis
(2) Quantitative Analysis
The combination of various mass spectrometry (MS) techniques, such as Liquid Chromatography-Mass Spectrometry (LC-MS) and Inductively Coupled Plasma-Mass Spectrometry (ICP-MS), is an effective approach for quantitative analysis of PCs. Employing ICP-MS allows for the exploration of the protein layer composition constituting the "hard protein coronas (HC)". Additionally, MS combined with chemical cross-linking can determine the binding sites between proteins within the PC. X-ray Photoelectron Spectroscopy (XPS) techniques enable the quantitative measurement of surface components of NPs. Through this method, the number of proteins on NPs and the dry thickness of protein layers can be estimated. The kinetic and thermodynamic constants during the formation of PC can be characterized using Fluorescence Correlation Spectroscopy (FCS). However, a drawback of this technique lies in its insensitivity to protein conformation. Fluorescence Spectroscopy/Tryptophan Fluorescence (FS-Trp) offers another method for quantitative analysis of PCs.
(3) Structural Analysis
The adsorption of proteins and structural changes in PCs can be detected using various techniques such as Fourier Transform Infrared Analysis (FTIR), X-ray Crystallography (XRD), Nuclear Magnetic Resonance (NMR), Raman Spectroscopy, Circular Dichroism (CD) Spectroscopy, Site-Directed Spin Labeling Electron Paramagnetic Resonance (SDSL-EPR), and Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS). CD monitoring can track changes in secondary structure during the interaction between PCs and NPs, providing insights into their binding mechanisms. FTIR spectroscopy is not limited by sample physical state or size. The atomic structure of PCs can be analyzed using X-ray crystallography, though it necessitates the preparation of large diffraction-quality samples. NMR provides high-resolution information on PCs in aqueous solutions under physiological conditions yet is limited by protein size. FTIR and Raman spectroscopy are useful for studying adsorbed protein secondary structures, while other spectroscopic techniques offer more detailed information on protein molecular structures. SDSL-EPR is an advanced technique for quantitative and qualitative analysis of adsorbed biomolecules. FS aids in determining binding constants. Dynamic nonspecific interactions between proteins and NPs can be studied using Surface Plasmon Resonance (SPR) sensors. Interaction between proteins and NPs can also be independently characterized using Isothermal Titration Calorimetry (ITC) and Fluorescence (FL) techniques, providing similar thermodynamic parameters such as binding constants (K) and Gibbs free energy (ΔG). UV, UV-Vis spectroscopy, and zeta potential analysis are alternative methods available.
(4) Protein Adsorption Analysis
Analytical techniques that explore protein-NP interactions are similarly useful for examining protein adsorption on NP surfaces. These approaches facilitate the measurement of adsorbed protein quantity, adsorption dynamics, and protein structural alterations. The selection of an appropriate method is guided by the protein's characteristics. UV-Vis spectrophotometry stands out as one of the favored techniques for quickly measuring adsorbed protein mass. Nonetheless, this analysis typically necessitates supplementary colorimetric analysis. Determining the change in protein mass in the solution upon NP surface contact is crucial, as it might influence result accuracy. Direct methods such as ToF-SIMS and SDSL-EPR are also viable for quantifying surface-adsorbed proteins. In the realm of adsorption dynamics, Quartz Crystal Microbalance (QCM) and Surface Plasmon Resonance (SPR) offer real-time, in-situ insights. Given the complexity of protein adsorption on NP surfaces, acquiring comprehensive information is essential for a full understanding.
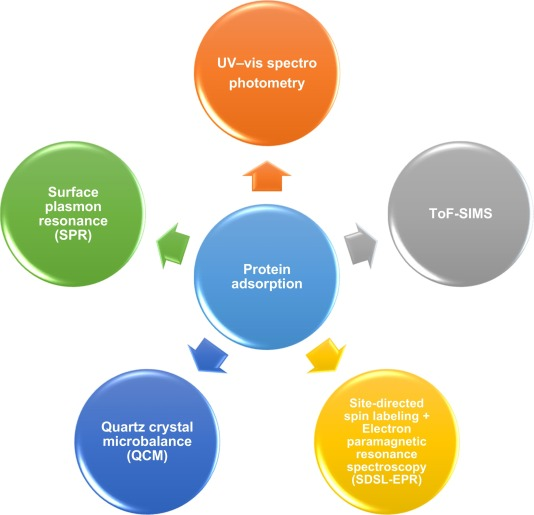
Figure 4. Methods for Analyzing PC Protein Adsorption[4]
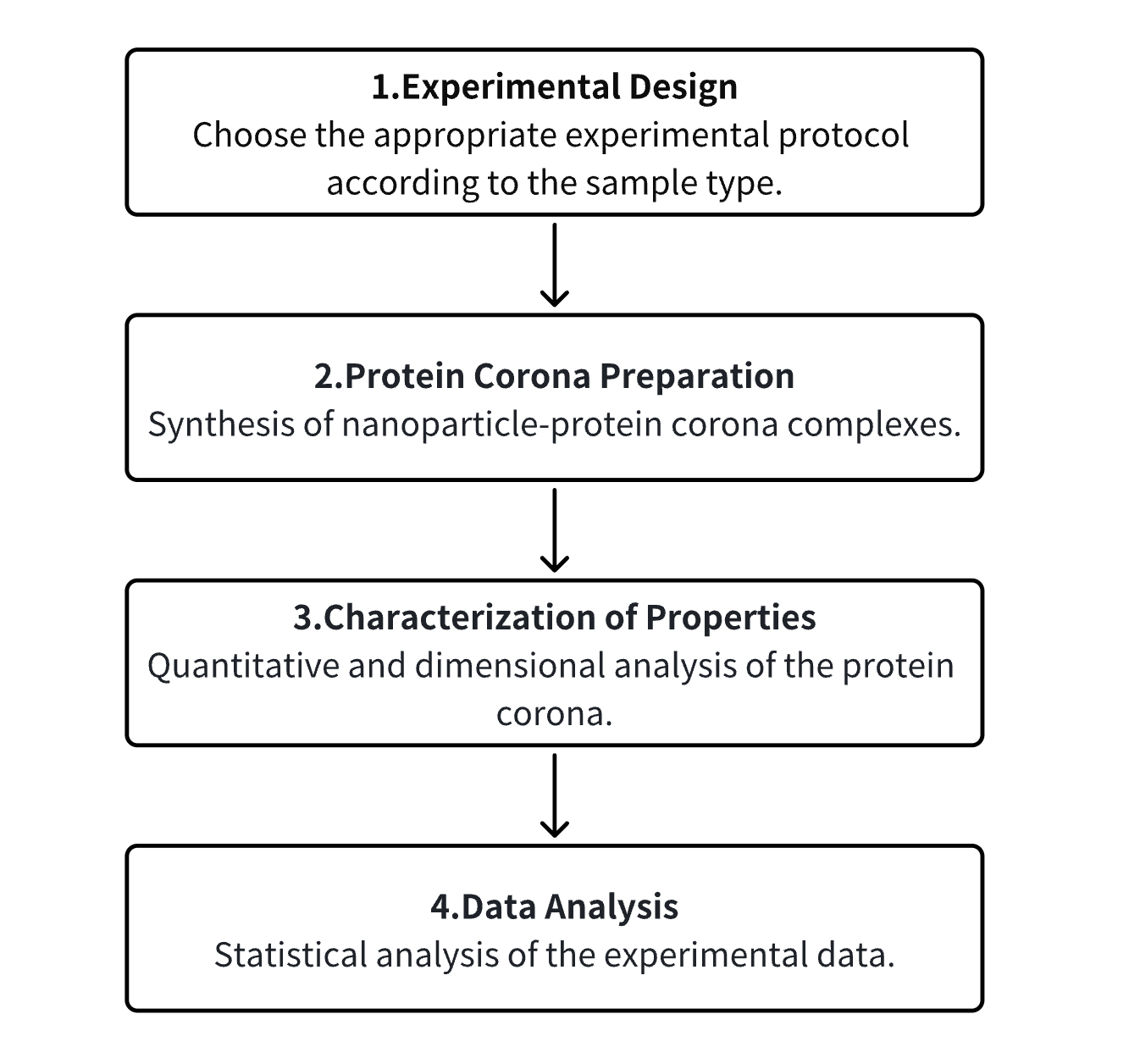
Analysis Workflow
1. Determine the Experimental Procedure Based on the Requirements of the Experiment
2. Preparation of the PC
3. Characterization of the PC's Various Properties
4. Data Analysis
Service Advantages
1. Characterization Services Offered for Diverse Properties of the PC
2. High Reliability and Precision in Detection
3. Comprehensive Bioinformatics Analyses
Sample Results
1. Localization and Identification of PCs on NP Surfaces and Their Impact on Cell Binding
Understanding the biological properties that NPs may acquire in a given biological environment is currently inferred from HC. Due to the lack of analytical separation methods, the composition of proteins in soft protein coronas (SC) and their biological relevance remain elusive. Studies have identified a set of specific PC proteins with weak interactions on silica and polystyrene NPs using in situ click chemistry. It was found that these "SC" proteins also exist in "HC", but are specifically enriched after capture, indicating that the main difference between HC and SC lies in the different binding strengths of the same proteins. Interestingly, proteins with weak interactions were primarily revealed as modulators of NP-cell binding through their dynamic properties. Therefore, the study emphasizes the consideration of protein-protein weak interactions on NPs when evaluating nano-bio interfaces.
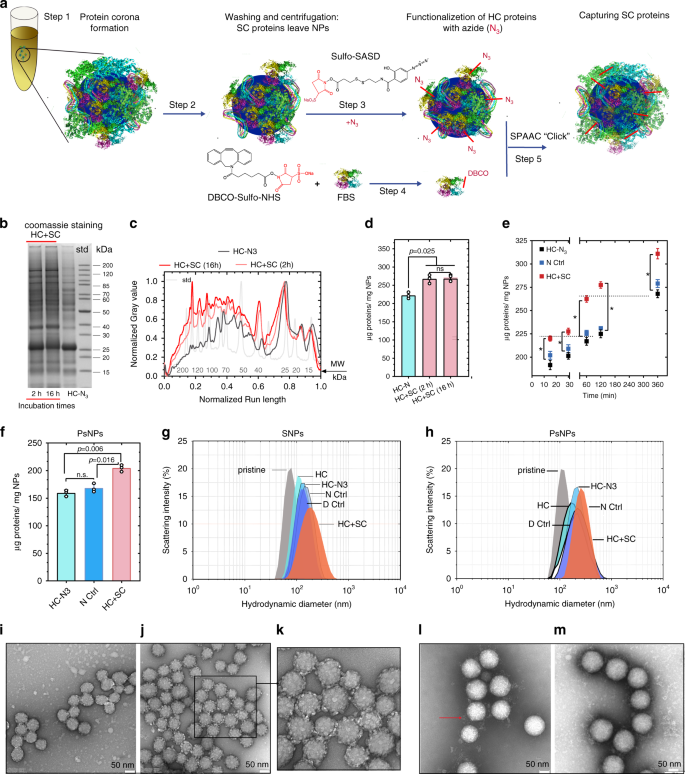
Figure 5. Characterization of SPAAC Click Chemistry Reactions and NP-PC Complexes[5]
2. Dynamic Heterogeneity of NP-PC Revealed by Super-Resolution Microscopy
The adsorption of serum proteins leads to the formation of biological molecule PCs, which are critical determinants of the in vivo biological properties of NPs. Therefore, understanding the formation, composition, and temporal evolution of PCs is crucial for the development of NP-based therapies. This study demonstrates that super-resolution optical microscopy can image PCs on mesoporous silica NPs with sensitivity to individual proteins. Particle-wise quantification reveals significant heterogeneity in protein absorption under native conditions. Furthermore, the diversity of coronas changes over time depending on the surface chemistry and degradation of particles. This study investigates the impact of protein adsorption on antibody-functionalized NP-specific cell targeting and provides detailed insights into the relationship between PCs and activity. This method is widely applicable to various nanostructures and complements existing approaches to PC research.
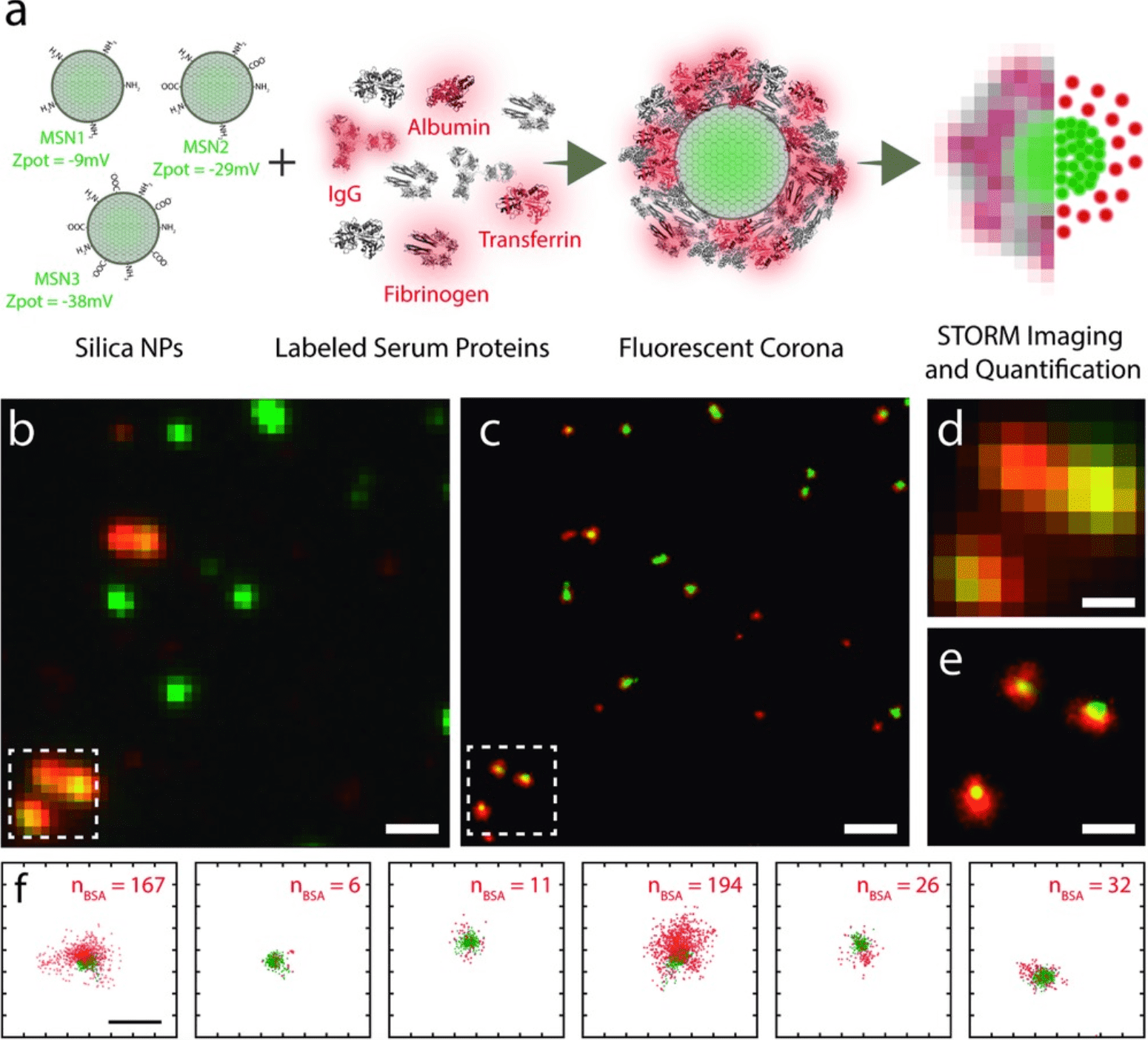
Figure 6. STORM Imaging of PC Formation in Silica NPs[6]
3. Formation of PC on the Surface of Extracellular Vesicles in Plasma
Studies have tested whether PCs form around extracellular vesicles (EVs) in plasma. Medium-sized nascent EVs were isolated from THP1 cells and Optiprep-purified platelets and subjected to MS analysis after differential centrifugation, size-exclusion chromatography, or density gradient ultracentrifugation. Compared to nascent EVs, plasma protein-coated EVs had higher densities and carried a large number of new associated proteins. Confocal microscopy, capillary Western blotting, immunoelectron microscopy, and flow cytometry confirmed interactions between plasma proteins and EVs.
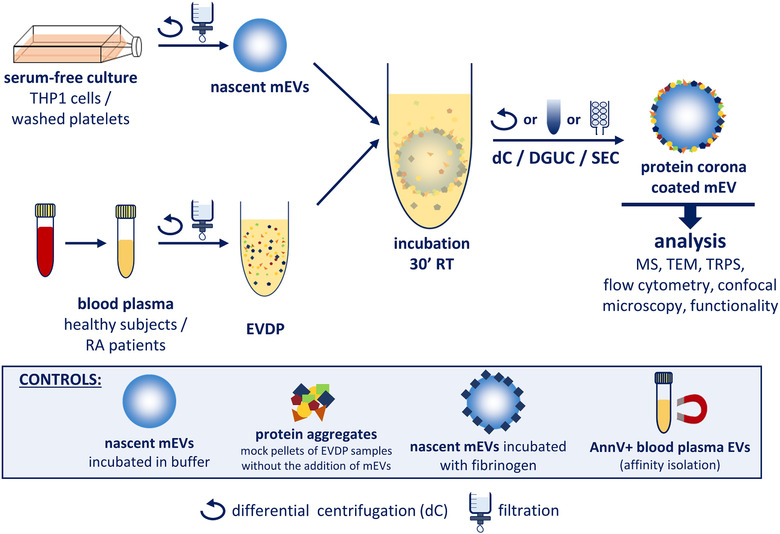
Figure 7. Experimental Workflow for EV PC Characterization[7]
Nine shared coronal proteins (Apolipoprotein A1, Apolipoprotein B, Apolipoprotein C3, Apolipoprotein E, Complement factors 3 and 4B, Fibrinogen alpha chain, Immunoglobulin heavy constant gamma 2 and gamma 4 chains) were identified, which seem to be shared coronal proteins of EVs in plasma, viruses, and artificial NPs. An unexpected finding of this study is that the composition of PCs highly overlaps with that of plasma protein aggregates. Besides diffuse, patchy PCs, large protein aggregates also associate with EV surfaces. However, while EVs loaded with external plasma protein cargo induced human monocyte-derived dendritic cells to express more TNF-α, IL-6, CD83, CD86, and HLA-DR, protein aggregates devoid of EVs had no effect.
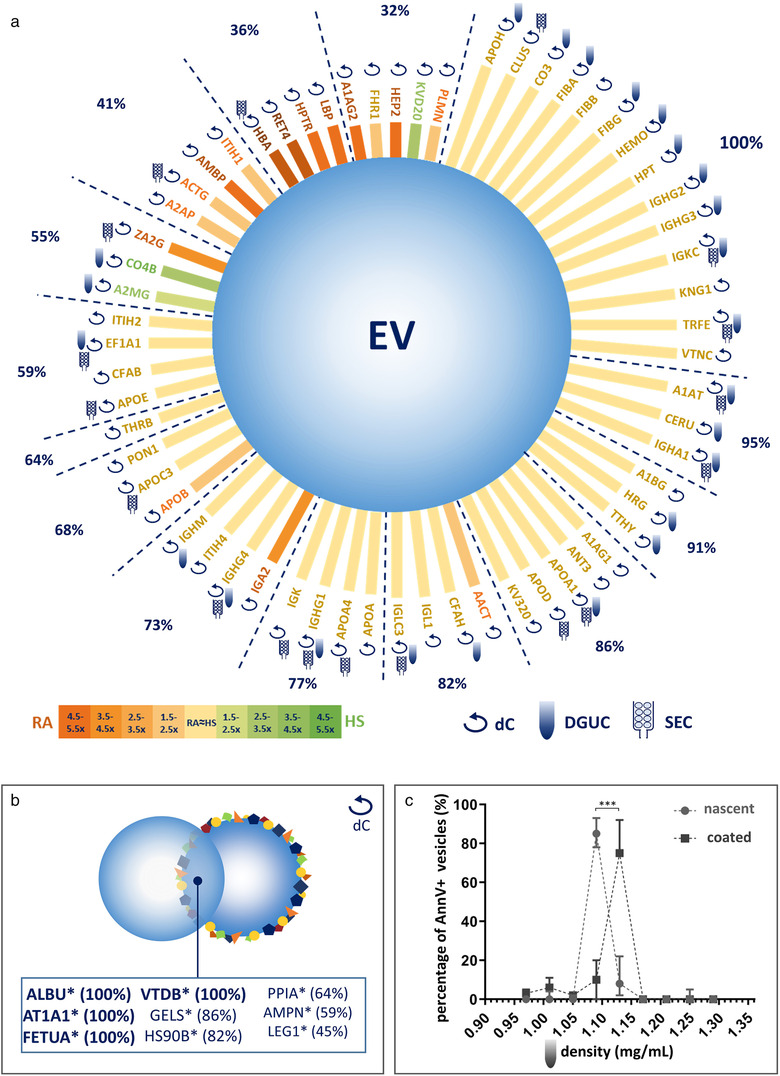
Figure 8. Proteomic Composition of the EV PCs [7]
Sample Submission Requirements
1. Efforts should be made to minimize contamination with impurities.
Services at MtoZ Biolabs
1. Comprehensive Experimental Procedures
2. Detailed Instrument Parameters
3. Raw Experimental Data
4. Data Analysis Report
Applications
1. Enhanced Permeability and Retention Nanotherapy
Chemotherapy is the standard treatment option in clinical oncology but often suffers from severe side effects and the development of resistance. One major factor is the unique physiological features of tumors that hinder the uniform distribution of low molecular weight (LMW) molecules, especially non-targeted chemotherapeutics, in the central regions of tumors. When these LMW molecules encounter high interstitial pressure (due to vascular heterogeneity caused by abnormal tumor microvasculature and inadequate lymphatic drainage within tumors) and rapid growth of tumor cells around blood vessels exceeding the diffusion limit, it reduces the local availability of drugs. In contrast, compared to LMW molecules, nanomedicines can prolong circulation and enhance accumulation within tumors via the Enhanced Permeability and Retention (EPR) effect. However, passive targeting of nanomedicines promoted by EPR shows highly variable responses in human clinical trials. Given the increasing consensus on extensive differences between and within patients, identifying patients with higher tumor-targeting potential through EPR would have significant benefits.
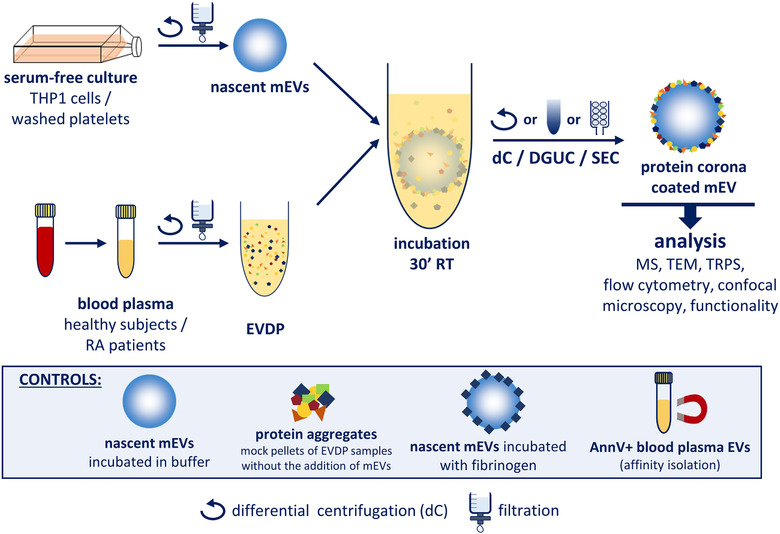
Figure 9. EPR Nanotherapy [2]
2. Development of Novel Technologies
Although the formation of PCs on NP surfaces typically leads to some adverse consequences (such as incorrect targeting in nanomedicine and triggering off-target responses), it also presents new opportunities for addressing issues ranging from early disease detection to ecological concerns. NPs can be specifically decorated to enhance targeting, for example, with immunostimulatory proteins, which can be used to modulate the immune system and catalyze the design of new therapies. Proteomic analysis of PCs on various NPs (e.g., sensor arrays) provides a unique opportunity to identify novel biomolecular patterns with disease detection capabilities. When NPs enter ecological environments, ecological coronas form, leading to spontaneous adsorption of proteins from ecological sources.
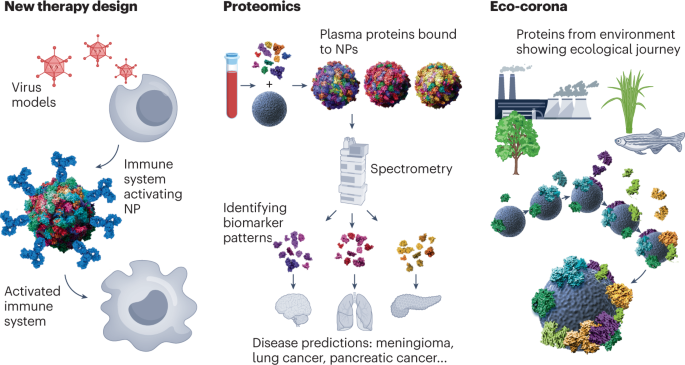
Figure 10. Emerging Technologies Based on PCs[8]
References
[1] Kim W, Ly NK, He Y, Li Y, Yuan Z, Yeo Y. PC: Friend or foe? Co-opting serum proteins for nanoparticle delivery. Adv Drug Deliv Rev. 2023 Jan;192:114635. doi: 10.1016/j.addr.2022.114635. Epub 2022 Nov 26. PMID: 36503885; PMCID: PMC9812987.
[2] Liu Y, Wang J, Xiong Q, Hornburg D, Tao W, Farokhzad OC. Nano-Bio Interactions in Cancer: From Therapeutics Delivery to Early Detection. Acc Chem Res. 2021 Jan 19;54(2):291-301. doi: 10.1021/acs.accounts.0c00413. Epub 2020 Nov 12. PMID: 33180454.
[3] Wheeler KE, Chetwynd AJ, Fahy KM, Hong BS, Tochihuitl JA, Foster LA, Lynch I. Environmental dimensions of the Protein Corona. Nat Nanotechnol. 2021 Jun;16(6):617-629. doi: 10.1038/s41565-021-00924-1. Epub 2021 Jun 11. PMID: 34117462.
[4] Kopac T. Protein Corona, understanding the nanoparticle-protein interactions and future perspectives: A critical review. Int J Biol Macromol. 2021 Feb 1;169:290-301. doi: 10.1016/j.ijbiomac.2020.12.108. Epub 2020 Dec 21. PMID: 33340622.
[5] Mohammad-Beigi H, Hayashi Y, Zeuthen CM, Eskandari H, Scavenius C, Juul-Madsen K, Vorup-Jensen T, Enghild JJ, Sutherland DS. Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat Commun. 2020 Sep 10;11(1):4535. doi: 10.1038/s41467-020-18237-7. PMID: 32913217; PMCID: PMC7484794.
[6] Feiner-Gracia N, Beck M, Pujals S, Tosi S, Mandal T, Buske C, Linden M, Albertazzi L. Super-Resolution Microscopy Unveils Dynamic Heterogeneities in Nanoparticle Protein Corona. Small. 2017 Nov;13(41). doi: 10.1002/smll.201701631. Epub 2017 Sep 18. PMID: 28922574.
[7] Tóth EÁ, Turiák L, Visnovitz T, Cserép C, Mázló A, Sódar BW, Försönits AI, Petővári G, Sebestyén A, Komlósi Z, Drahos L, Kittel Á, Nagy G, Bácsi A, Dénes Á, Gho YS, Szabó-Taylor KÉ, Buzás EI. Formation of a protein corona on the surface of extracellular vesicles in blood plasma. J Extracell Vesicles. 2021 Sep;10(11):e12140. doi: 10.1002/jev2.12140. PMID: 34520123; PMCID: PMC8439280.
[8] Mahmoudi, M., Landry, M.P., Moore, A. et al. The protein corona from nanomedicine to environmental science. Nat Rev Mater 8, 422–438 (2023). https://doi.org/10.1038/s41578-023-00552-2
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
MS-Based Protein Corona Characterization Service | Nanoparticles
Protein Corona-Mediated Nanomedicine Targeted Delivery Service
How to order?







