MS-Based Protein Corona Characterization Service | Nanoparticles
The protein corona (PC) of nanoparticles (NPs) exhibits several advantageous properties. Firstly, it can selectively enrich or deplete specific proteins within the corona, independently of the protein composition present in biological fluids. This capability is particularly beneficial for protein identification and characterization, especially within the realm of plasma proteomics. Secondly, the composition of the PC on the same NPs can be influenced by the donor's disease type. This idea of disease-specific PCs has been corroborated by various research groups and applied in studying how these PCs, which are personalized and disease-specific, interact with biological systems. Additionally, although fewer proteins are detected in the PC compared to native biological fluids, the reduced number of plasma proteins could potentially increase the sensitivity and specificity of disease and biomarker detection.
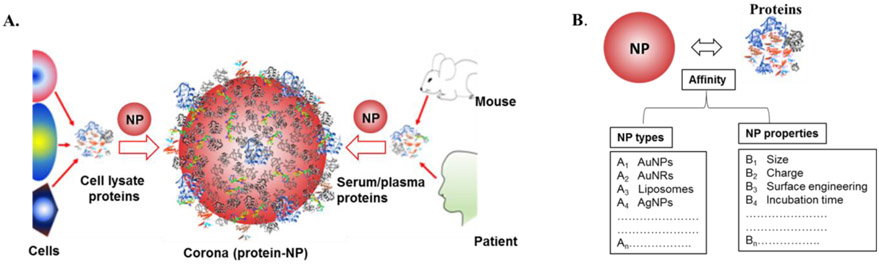
Figure 1. Nanoparticle-Protein Corona [1]
The majority of experimental procedures in PC research that use a bottom-up approach encompass five principal steps: exposing NPs to biological fluids (I), isolating NP-protein complexes (II), digesting the proteins of the PC (III), separating peptides (IV), and identifying peptides through MS/MS and bioinformatics (V).
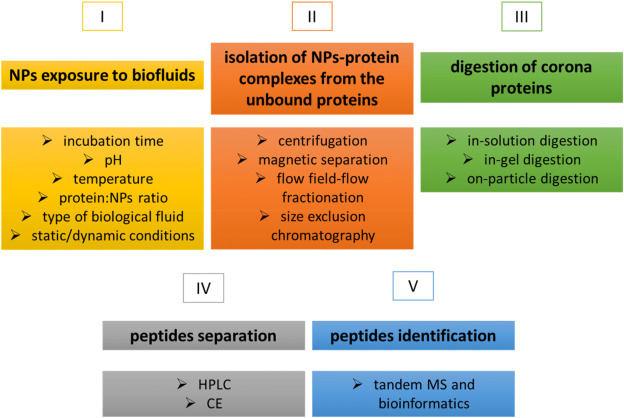
Figure 2. Protocol for PC Identification Using a Bottom-Up Approach[2]
1. Formation of the PC
The formation of the PC is influenced by several key factors, including incubation time, pH, temperature, the protein-to-NP ratio, and the type of biological fluid. The biological fluid used should mimic the internal body environment, with human plasma and serum being the most commonly used. Most in vitro studies, however, neglect that NPs injected intravenously in vivo are continuously mobile and interact with serum proteins and cell membrane components in a dynamic environment. Research has shown that compared to static incubation, liposomal NPs incubating under continuous flow conditions (simulating human abdominal aorta blood flow speeds) yield different PC compositions. Additionally, microfluidic devices have proven effective for the dynamic incubation of NPs with proteins, enhancing reproducibility across laboratories and facilitating standardization of incubation protocols.
Figure 3. Formation of the PC[1]
2. Separation of the PC
Reliable identification of the PC necessitates separating NP-protein complexes from the biological matrix without altering the corona's protein characteristics or disrupting protein equilibrium.
Centrifugation, the most commonly employed method, capitalizes on the density differences between the protein-crowned NPs and matrix proteins. Its primary benefits include ease of operation and ubiquitous equipment availability. However, centrifugation parameters (such as speed, duration, and washing conditions) must be meticulously optimized to prevent false positives—non-PC protein aggregates precipitating during centrifugation—and false negatives caused by PC detachment from NPs due to centrifugal forces.
Magnetic separation, used for NPs with superparamagnetic properties, minimizes structural impacts on complexes and reduces false positives associated with protein aggregation during centrifugation. Though this method requires washing, it minimizes analyte loss. In instances where magnetic separation alone is insufficient, additional centrifugation may be required to collect smaller NPs.
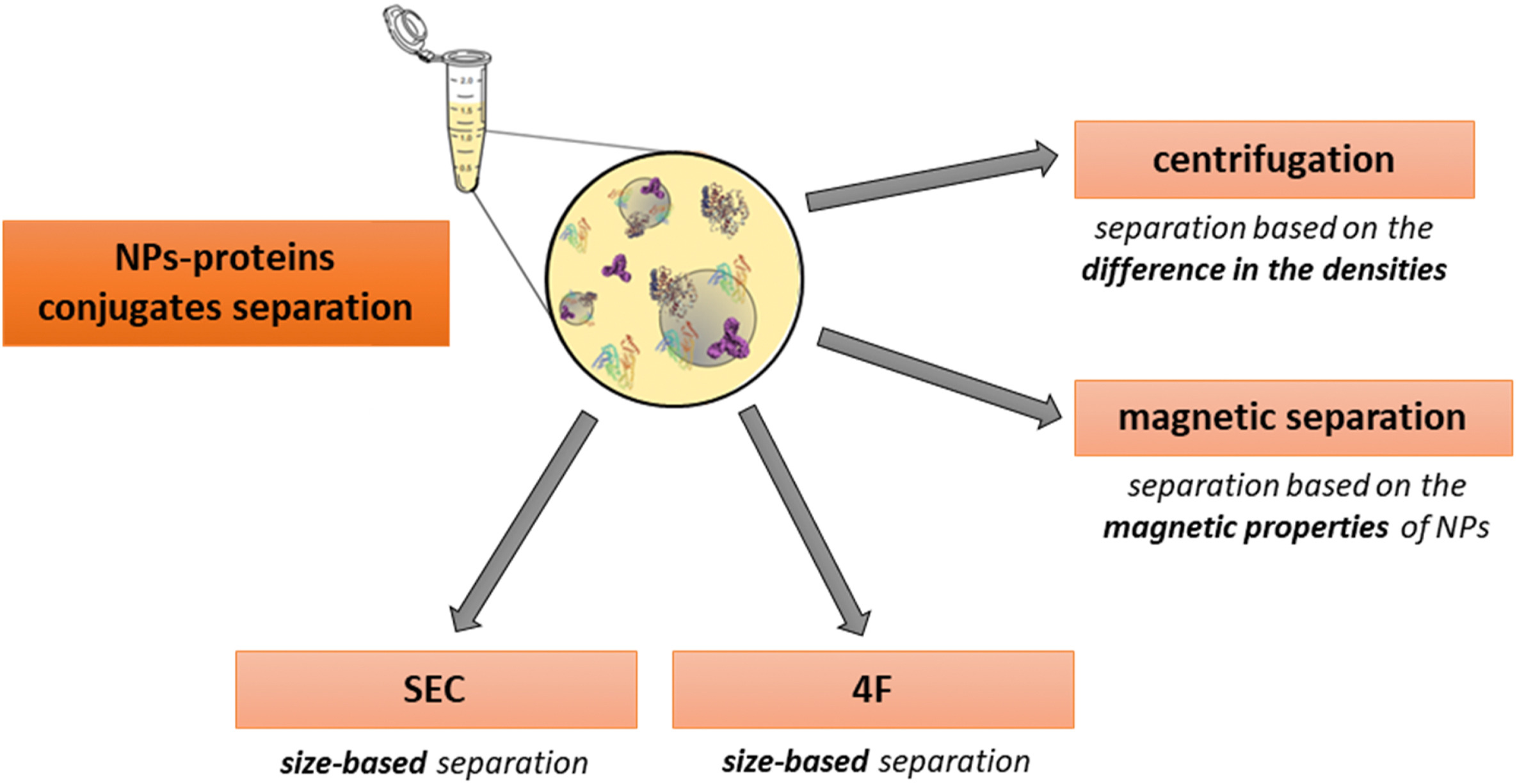
Figure 4. Separation of the PC[2]
NPs coated with a PC can be separated using Flow Field-Flow Fractionation (F4). This method, which does not use column packing, minimizes interactions with the NP-protein complexes, employing an open-channel technique. The F4 method isolates fractions based on the size of the complexes for further analysis. However, it has drawbacks, such as shear forces that may alter the PC, and the necessity for time-consuming parameter optimization. When used in conjunction with LC-MS/MS, F4 can identify slowly dissociating proteins from the PC.
Size Exclusion Chromatography (SEC) is another method that separates NP-protein complexes with minimal disturbance compared to centrifugation. SEC can isolate fractions and typically offers higher resolution than F4. However, potential interactions with the stationary phase may alter the PC. As an alternative, recent studies have introduced a microfluidic system-based separation method, where PC parameters are characterized by Multi-Parameter Surface Plasmon Resonance (MP-SPR), followed by LC-MS/MS analysis.
3. Enzymatic Digestion of the PC
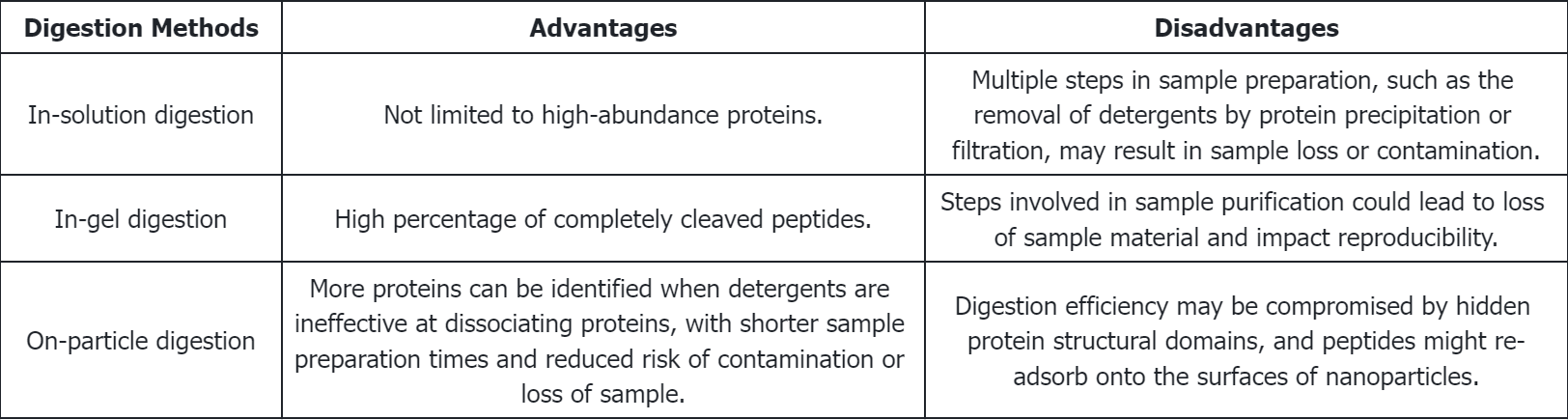
(1) In-Gel Digestion: Early research identifying PCs typically employed in-gel digestion procedures. This method separates proteins from NPs by brief high-temperature incubation in SDS-PAGE sample buffers containing surfactants. After separation by SDS-PAGE, individual protein bands are excised from the gel and digested. Peptides are then extracted from the gel using organic solvents for subsequent analysis.
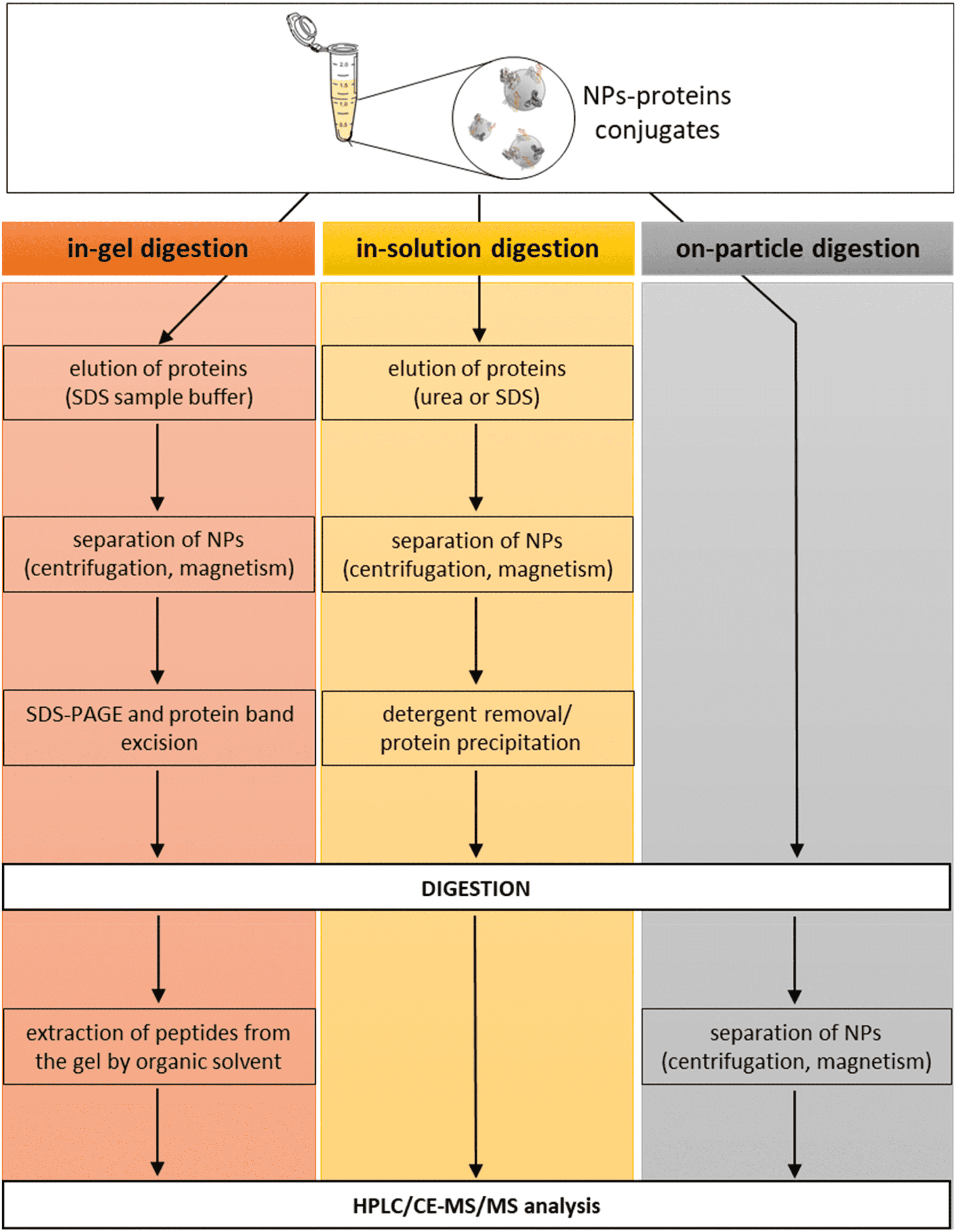
Figure 5. Enzymatic Digestion of the PC[2]
(2) In-Solution Digestion: Proteins are initially dissociated from NPs by denaturants (such as high concentrations of urea or guanidine hydrochloride) or surfactants (e.g., SDS, LDS, or CHAPS), followed by digestion in solution. Chelating agents like EDTA may also be employed. The dissociation of the PC is a critical step; incomplete dissociation can lead to inaccurate quantitative and qualitative results. Therefore, it is essential to choose appropriate dissociation agents and conditions. For instance, research indicates that SDS effectively removes most proteins from polystyrene NPs, whereas CHAPS selectively extracts lipoproteins.
(3) On-Particle Digestion: On-particle digestion serves as an alternative to solution and gel-based methods. This technique directly digests proteins adsorbed on NP surfaces and separates the resulting peptides from NPs through centrifugation or ultrafiltration. This approach has the advantage of requiring fewer procedural steps, thereby minimizing risks like protein loss or sample contamination. Moreover, this direct digestion process may handle more proteins than can be desorbed by detergents from NP surfaces. Additionally, shortening the sample preparation enhances reproducibility and accuracy. However, on-particle digestion has drawbacks: proteins may change conformation upon adsorption to NPs, or steric hindrance from adjacent proteins might prevent trypsin from accessing digestion sites. Additionally, peptides might re-adsorb onto NP surfaces post-digestion, complicating further identification. Strategies to mitigate these challenges include deactivating the NP surface or modifying solution conditions to lessen peptide re-adsorption.
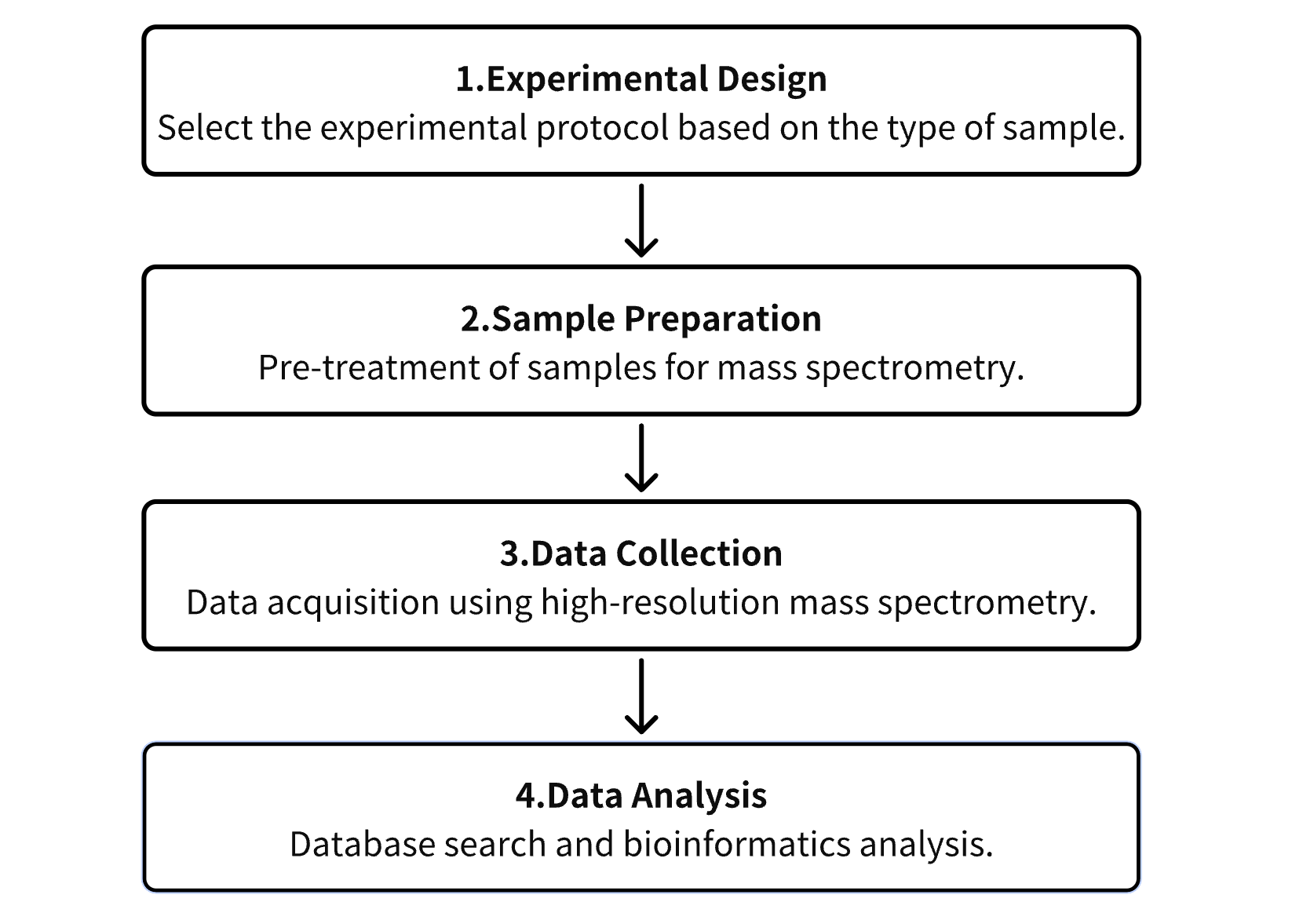
Analysis Workflow
1. Establish the Experimental Protocol Based on Specific Experimental Objectives
2. Preparation of Samples for Mass Spectrometry (MS) Analysis
3. Collection of Data Using High-Resolution MS
4. Retrieval and Analysis of Experimental Data
Service Advantages
1. Identification, Quantification, and Characterization of Protein Modifications from Diverse Sample Sources
2. High-Confidence, High-Precision MS Analysis
3. Comprehensive Bioinformatics Analysis
Sample Results
1. Rapid, In-Depth, and Precise Analysis of the Plasma Proteome Using Multi-NPs
Researchers have identified large-scale, unbiased proteomic studies as being constrained by the complexity of the plasma proteome. A recent publication describes a highly parallel protein quantification platform that integrates NP PCs with liquid chromatography-mass spectrometry (LC-MS). This combination enhances efficient proteomic analysis. The PC, a layer of proteins adsorbed onto NPs upon exposure to biological fluids. By modifying the physicochemical properties of engineered NPs, researchers can achieve distinct PC patterns, enabling differentiated and reproducible detection. Experimental results have demonstrated a linear signal response with a median coefficient of variation of 22%. In classification trials for non-small cell lung cancer, a 96-well automated workflow screened 43 types of NPs and identified 5 NPs capable of detecting more than 2000 proteins in 141 plasma samples. This streamlined workflow, which integrates depth of coverage and throughput with precise quantification based on unique interactions between proteins and NPs, supports extensive and scalable quantitative proteomics research.
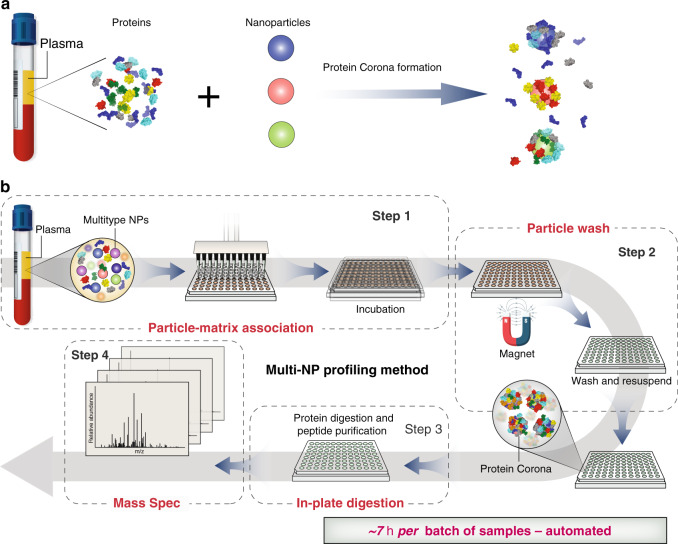
Figure 6. Workflow for Plasma Proteome Analysis Using Multi-NP PC [3]
2. Using Multi-NP to Analyze the Human Plasma Proteome for Alzheimer's Disease Detection
As the prevalence of Alzheimer's Disease (AD) increases, the need for non-invasive and accurate diagnostic tools also grows. Current research indicates that AD pathogenesis begins decades before the onset of clinical symptoms. NPs possess unique properties that make them suitable for developing early detection diagnostics for AD. Upon interacting with biological fluids, NPs undergo unbiased but selective and reproducible adsorption of biomolecules, forming what is known as the PC. The discovery of differential changes in the plasma proteome during health and disease has led to the concept of disease-specific PCs. Using multi-NP platforms, researchers have successfully identified subtle variations in plasma protein patterns and detected AD with over 92% specificity and approximately 100% sensitivity. This approach has proven effective in discerning AD from plasma samples collected years before diagnosis. The platforms' ability to analyze pathological proteomic changes as disease-specific markers underscores the significance of this promising non-invasive technique for early detection and intervention, benefiting not only AD patients but potentially those with other diseases as well.
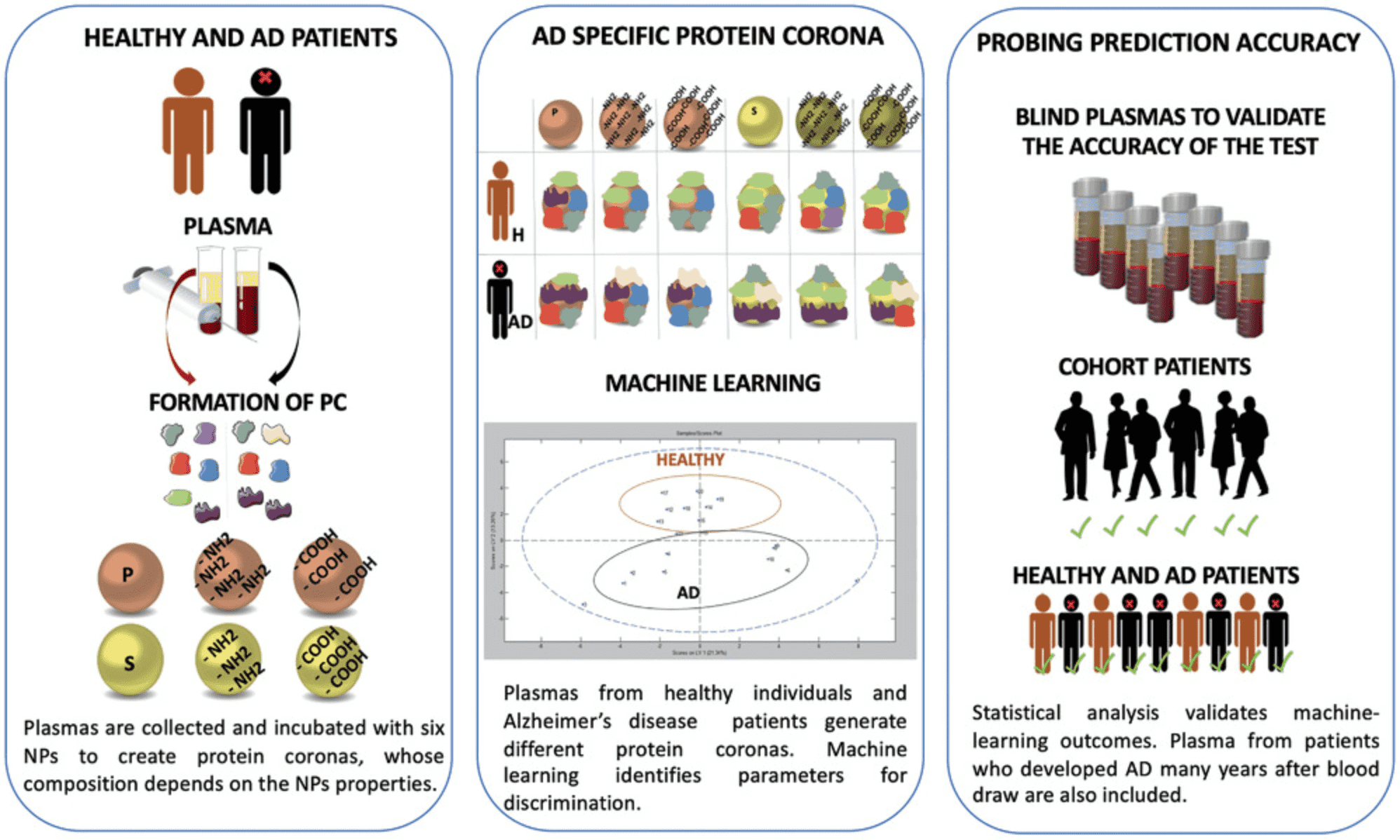
Figure 7. Analyzing Alzheimer's Disease Plasma Proteome Using Multi-NP PC [4]
3. Proteomics Reveals Time-Dependent Changes in PCs Within Intracellular Pathways
The dynamics and formation of intracellular PCs remain underexplored in the nano-bio interaction domain. Researchers have established a workflow to isolate intracellular PCs of two types—magnetic hydroxyethyl starch nanoparticles (HES-NPs) and magnetic human serum albumin nanocapsules (HSA-NCs)—at varying uptake times. The study employed label-free quantitative LC-MS proteomics to analyze the composition of the intracellular PCs, correlating these findings with traditional transport methods like flow cytometry, transmission electron microscopy (TEM), and confocal laser scanning microscopy (cLSM). The study highlights the evolutionary patterns of intracellular PCs; HES-NPs exhibited significant changes over time, while HSA-NCs displayed less variability. The study identified proteins involved in macropinocytosis (RAC1, ASAP2) as well as clathrin, with further confirmation through blocking experiments and TEM studies. The study's nanocarriers were primarily identified from early endosomes to late endosomes/lysosomes (RAB7, LAMP1, cathepsin K, and HSP 90-β) via proteomics and cLSM through RAB5. This research differentiates between NPs with slow and fast uptake dynamics, identifying relevant proteomic profiles at various time points, thereby providing valuable data for efficient drug delivery and intracellular applications using NPs.
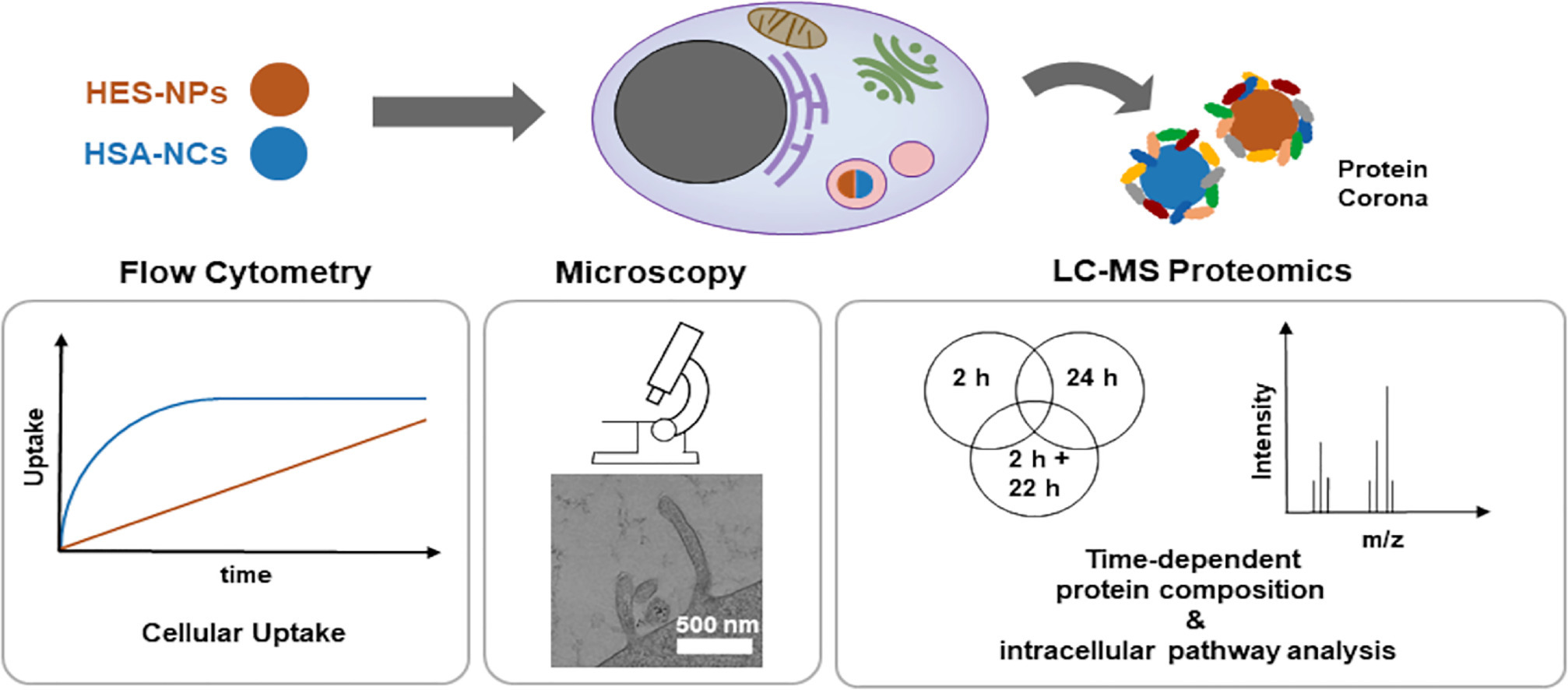
Figure 8. Intracellular PC Analysis Using Multi-NP PC [5]
Sample Submission Requirements
1. Protein Purity >90%
2. Minimize the Risk of Contamination with Impurities
Services at MtoZ Biolabs
1. Complete Experimental Steps
2. Instrument-Related Parameters
3. Original Experimental Data
4. Data Analysis Report
Applications
1. Personalized PC
Factors such as gender, habits, age, disease, drug intake, pregnancy, and geographic origin can influence the composition of blood plasma proteins, which subsequently alters the PC composition. Consequently, analyzing the PC can facilitate the diagnosis of specific pathological states. Various carriers serve as enrichment platforms, enabling the analysis of differences in human plasma proteomes under different conditions.
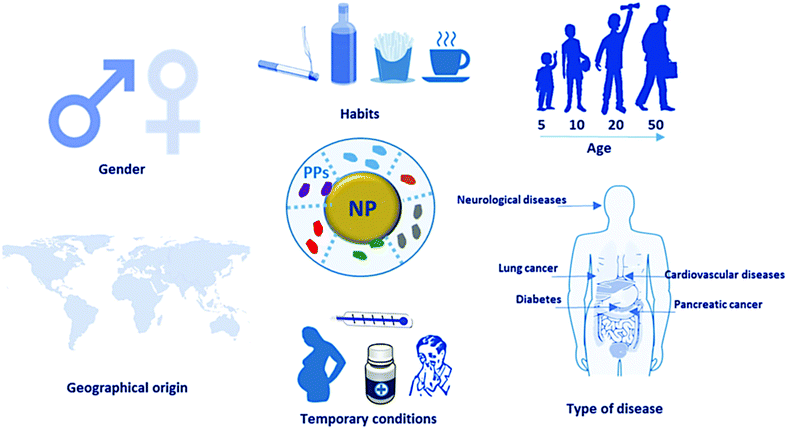
Figure 9. General Concept of Personalized PC [6]
2. Development of New Materials for PC Research
In vitro diagnostics (IVD) constitute a crucial component of medical examinations, increasingly vital in clinical care. Omics studies are poised to enhance diagnostic precision and provide a more comprehensive assessment of diseases, demonstrating substantial potential for application in IVD. Nonetheless, the omics analysis of biological fluids faces several challenges due to the high complexity of samples and low molecular abundance. Advances in technology, featuring diverse morphologies and compositions, facilitate the separation, enrichment, and ionization of disease-associated biomolecules in omics studies. Currently, nanomaterials optimized for omics analysis offer advantages such as high throughput, straightforward pre-treatment, and rapid detection, and have found extensive applications in IVD.
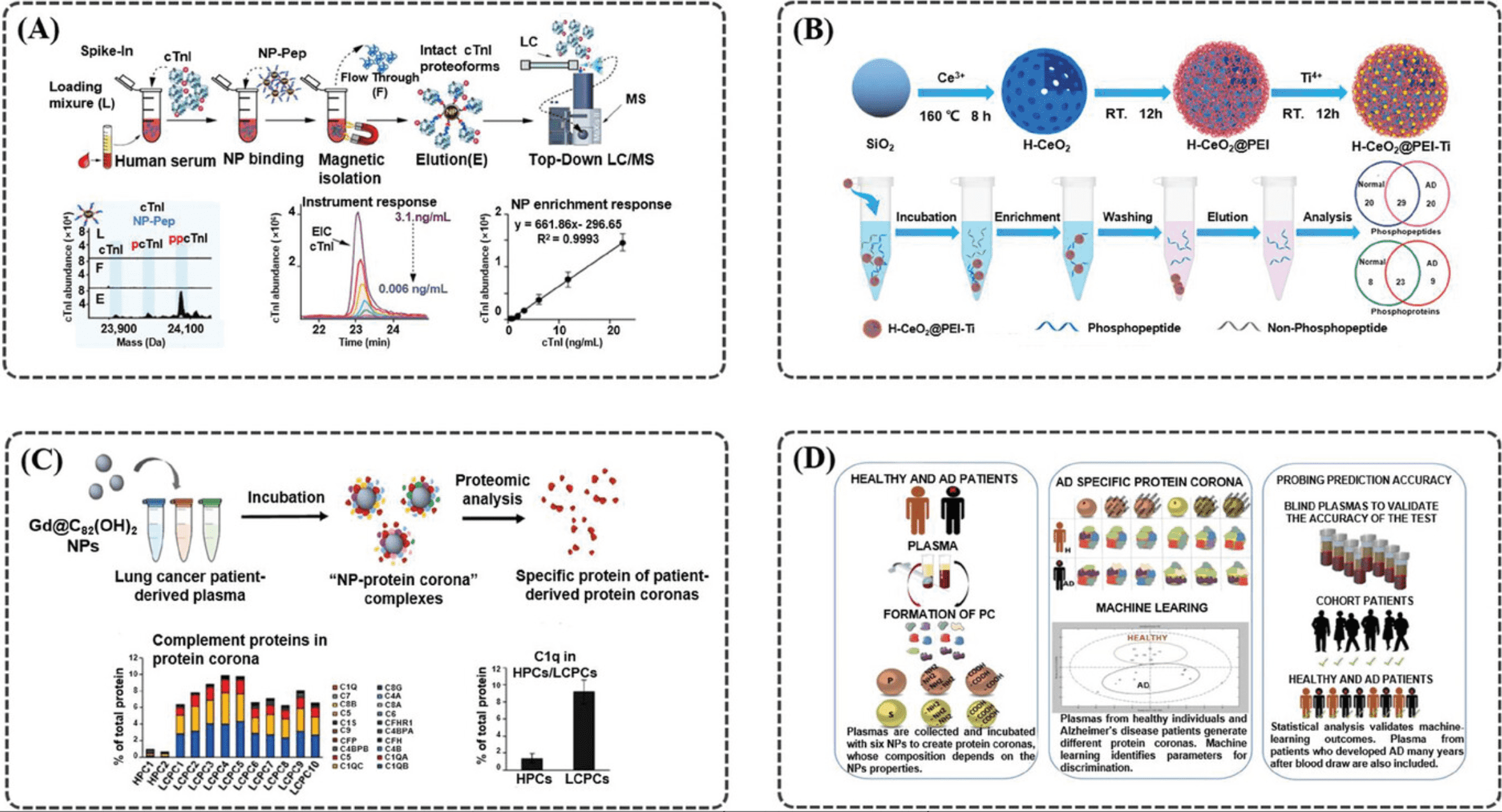
Figure 10. Different Materials Used for Proteomics Analysis [7]
FAQ
Q1: What are the respective advantages and disadvantages of various enzymatic digestion methods?
References
[1] Elechalawar CK, Hossen MN, McNally L, Bhattacharya R, Mukherjee P. Analysing the nanoparticle-protein corona for potential molecular target identification. J Control Release. 2020 Jun 10;322:122-136. doi: 10.1016/j.jconrel.2020.03.008. Epub 2020 Mar 9. PMID: 32165239; PMCID: PMC7675788.
[2] Kruszewska J, Zajda J, Matczuk M. How to effectively prepare a sample for bottom-up proteomic analysis of nanoparticle protein corona? A critical review. Talanta. 2021 May 1;226:122153. doi: 10.1016/j.talanta.2021.122153. Epub 2021 Jan 29. PMID: 33676702.
[3] Blume JE, Manning WC, Troiano G, Hornburg D, Figa M, Hesterberg L, Platt TL, Zhao X, Cuaresma RA, Everley PA, Ko M, Liou H, Mahoney M, Ferdosi S, Elgierari EM, Stolarczyk C, Tangeysh B, Xia H, Benz R, Siddiqui A, Carr SA, Ma P, Langer R, Farias V, Farokhzad OC. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat Commun. 2020 Jul 22;11(1):3662. doi: 10.1038/s41467-020-17033-7. PMID: 32699280; PMCID: PMC7376165.
[4] Corbo C, Li AA, Poustchi H, Lee GY, Stacks S, Molinaro R, Ma P, Platt T, Behzadi S, Langer R, Farias V, Farokhzad OC. Analysis of the Human Plasma Proteome Using Multi-Nanoparticle Protein Corona for Detection of Alzheimer's Disease. Adv Healthc Mater. 2021 Jan;10(2):e2000948. doi: 10.1002/adhm.202000948. Epub 2020 Nov 9. PMID: 33169521.
[5] da Costa Marques R, Hüppe N, Speth KR, Oberländer J, Lieberwirth I, Landfester K, Mailänder V. Proteomics reveals time-dependent protein corona changes in the intracellular pathway. Acta Biomater. 2023 Dec;172:355-368. doi: 10.1016/j.actbio.2023.10.010. Epub 2023 Oct 13. PMID: 37839632.
[6] Kamaly N, Farokhzad OC, Corbo C. Nanoparticle protein corona evolution: from biological impact to biomarker discovery. Nanoscale. 2022 Feb 3;14(5):1606-1620. doi: 10.1039/d1nr06580g. PMID: 35076049.
[7] Wang Y, Li R, Shu W, Chen X, Lin Y, Wan J. Designed Nanomaterials-Assisted Proteomics and Metabolomics Analysis for In Vitro Diagnosis. Small Methods. 2023 Nov 3:e2301192. doi: 10.1002/smtd.202301192. Epub ahead of print. PMID: 37922520.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Protein Corona Characterization Service
Protein Corona-Mediated Nanomedicine Targeted Delivery Service
How to order?







