4D Proteomics Service
- Improved Qualitative Accuracy: Ion mobility separation allows for effective differentiation of modification isomers, leading to more precise identification of protein modifications.
- Increased Protein Identification Capacity: 4D proteomics enhances detection throughput by up to tenfold compared to conventional methods, thereby significantly shortening experimental timelines.
- Enhanced Quantitative Precision: The superior stability of the instrumentation achieves a correlation coefficient of over 0.95 for sample injections ranging from 1 to 96.
- Greater Sensitivity: Traditional proteomics typically requires microgram-level samples, whereas 4D proteomics can detect proteins at amounts as low as 200 ng, one-tenth the size of standard samples, making it ideal for analyzing trace samples.
- Profiling protein expression related to drug responses and gene knockout/overexpression studies
- Investigating the physiological processes underlying plant and microbial growth, development, and responses to environmental stress
- Conducting high-throughput analyses of disease biomarkers and drug targets
- Elucidating protein mechanisms and screening for proteins with unique biological functions
- Developing comprehensive proteomic maps across various species
The fundamental principle of MS-based proteomics is primarily analyzing according to the physicochemical properties of the sample ions, obtaining qualitative and quantitative results based on the sample's mass spectrum and related information. Currently, proteomics mass spectrometry generally identifies and quantifies peptides and proteins based on three dimensions: retention time (RT), mass-to-charge ratio (m/z), and ion intensity, named as 3D proteomics. 4D proteomics adds another dimension-ion mobility- on the basis of 3D proteomics, which separates ions mainly based on their shape and cross-section and can distinguish peptide segments with very small m/z differences to distinguish and recognize low-abundance protein signals. 4D proteomics, based on the timsTOF Pro mass spectrometer, combines parallel accumulation serial fragmentation (PASEF) with trapped ion mobility spectrometry (TIMS). It can repeatedly measure the collision cross-section (CCS) of all detected ions, enabling faster and more sensitive qualification and quantification of proteomics.
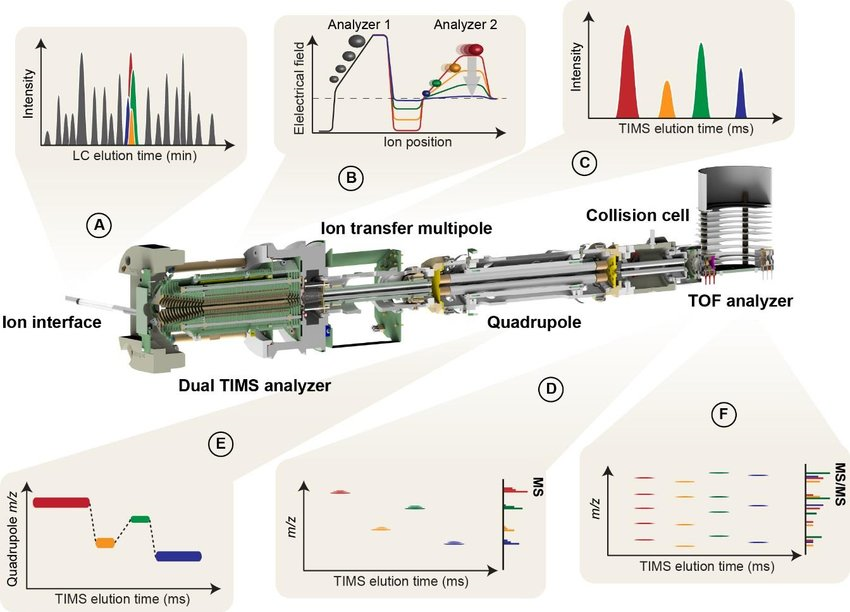
Meier, F. et al. Mol Cell Proteomics. 2018.
Figure 1. 4D Proteomics
MtoZ Biolabs provides 4D Proteomics Service based on timsTOF Pro, supporting proteomics research of trace samples, large medicine sample group, and high-throughput modificomics.
Service Advantages
Compared with traditional 3D proteomics, 4D proteomics offers several distinct benefits:
Applications
Sample Submission Requirements
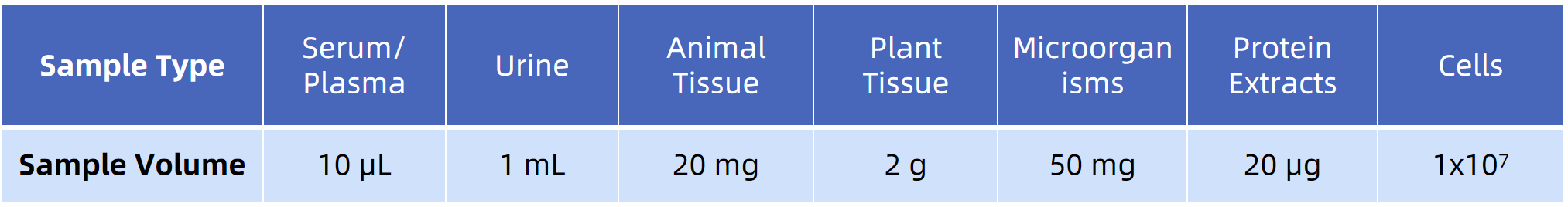
Note: Please use sufficient dry ice for transportation and choose a faster mailing method to reduce sample degradation during transportation.
Deliverables
1. Experimental Procedures
2. Relevant Experimental Parameters
3. Mass Spectrometry Images
4. Raw Data
5. Proteomics Analysis Results
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
4D Proteomics and Dual-Platform Full-Spectrum Metabolomics
How to order?







