Which Technologies Are Most Suitable for Spatial Proteomics Data Acquisition?
-
Visium Combined with Mass Spectrometry: Spatial RNA localization using 10x Genomics Visium, followed by region-specific protein analysis via LC-MS.
-
DSP (Digital Spatial Profiling): The NanoString platform enables simultaneous spatial profiling of both proteins and RNAs.
-
Optimized protocols for low-input protein extraction
-
Development of high-sensitivity LC-MS/MS methodologies
-
Customized antibody screening and labeling services for region-specific sampling
Spatial proteomics has emerged as a rapidly advancing frontier in life sciences, aiming to decipher the spatial distribution and dynamic alterations of proteins within tissues or cellular microenvironments. To fulfill this objective, data acquisition technologies must integrate high spatial resolution, robust protein detection throughput, and precise quantification. Currently, mainstream spatial proteomic acquisition approaches fall into two broad categories: imaging-based techniques and mass spectrometry-based techniques. Each offers unique advantages and is tailored to specific research applications.
Imaging-Based Spatial Proteomic Techniques
These methods typically employ antibody or probe labeling strategies to visualize and quantify protein localization through advanced imaging systems.
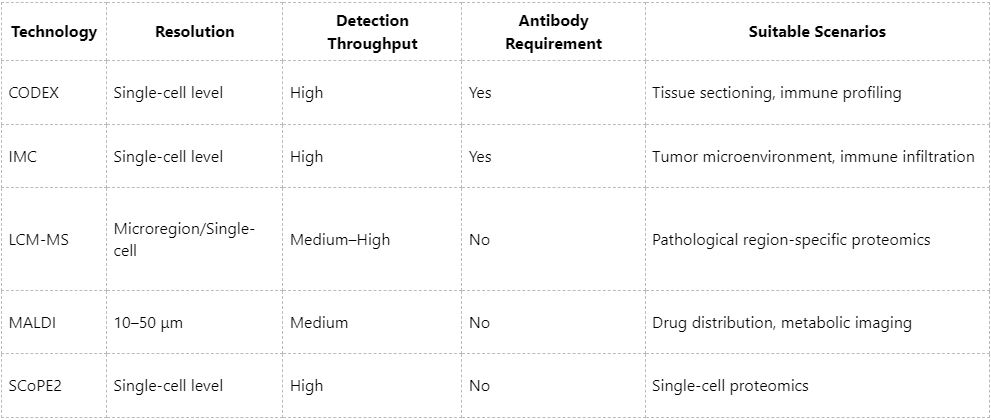
1. CODEX (CO-Detection by Indexing)
(1) Principle: Implements an antibody barcoding strategy, enabling detection of over 50 proteins through iterative rounds of fluorescence imaging.
(2) Advantages: High detection throughput and cellular-level spatial resolution; well-suited for tissue section analysis.
(3) Limitations: Dependent on high-quality antibodies; throughput constrained by optical system performance and cycle recognition efficiency.
2. MIBI (Multiplexed Ion Beam Imaging)
(1) Principle: Utilizes metal-tagged antibodies and secondary ion mass spectrometry (SIMS) for imaging-based protein detection.
(2) Advantages: Enables detection of over 40 protein targets with minimal fluorescence crosstalk and low background signal.
(3) Limitations: High instrumentation cost and relatively slow imaging speed; ideal for high-resolution histopathological studies.
3. Imaging Mass Cytometry (IMC)
(1) Principle: Integrates CyTOF with laser ablation to convert spatial signals from metal-labeled antibodies into mass spectrometry readouts.
(2) Advantages: Offers high multiplexing capacity and low antibody interference; suitable for spatial profiling of pathological tissues.
(3) Applications: Spatial characterization of tumor microenvironments, immune infiltration, and related studies.
Mass Spectrometry-Based Spatial Proteomic Techniques
These strategies segment tissue samples based on spatial coordinates via in situ digestion, laser microdissection, or spatial barcoding, followed by proteomic analysis using mass spectrometry.
1. LCM-MS (Laser Capture Microdissection + MS)
(1) Principle: Isolates specific tissue regions using laser microdissection for subsequent protein extraction and MS-based analysis.
(2) Advantages: Provides high spatial resolution (up to single-cell level) and deep proteome coverage.
(3) Challenges: Labor-intensive sample preparation and complex operations; best suited for targeted microregion investigations.
2. MALDI Imaging (Matrix-Assisted Laser Desorption/Ionization Imaging)
(1) Principle: Applies a matrix to the sample surface, followed by point-by-point laser scanning and TOF-MS to map protein distributions.
(2) Advantages: Antibody-free detection, high spatial resolution (~10 μm), applicable to both proteins and metabolites.
(3) Limitations: Restricted protein coverage; primarily used for spatial visualization rather than deep proteomic profiling.
3. SCoPE2 (Single-Cell Proteomics by Mass Spectrometry)
(1) Principle: Combines TMT labeling and nanoflow LC-MS/MS for single-cell level proteomic analysis.
(2) Advantages: High sensitivity and quantitative precision; suitable for analysis following spatial localization of single cells.
(3) Limitations: Requires complementary spatial localization strategies, such as microscopy or spatial barcoding.
Emerging Trend: Spatial Proteomics Integrated with Multi-Omics
Spatial proteomics is increasingly evolving toward multi-omics integration, in combination with spatial transcriptomics and metabolomics platforms, including:
MtoZ Biolabs' Technological Advantages in Spatial Proteomics
At MtoZ Biolabs, we have developed a comprehensive workflow for spatial proteome profiling in both FFPE and fresh tissue sections by integrating state-of-the-art laser capture microdissection systems (LCM) with high-resolution mass spectrometry platforms (e.g., Orbitrap Exploris series). Our innovations include:
These capabilities allow researchers to achieve both high spatial resolution and deep quantitative proteomic insights, applicable to investigations in tumor microenvironments, tissue heterogeneity, neurodevelopment, and more.
Summary: Matching Technologies to Research Needs
For more information on spatial proteomics experimental design, data analysis, or customized service solutions, please contact MtoZ Biolabs. We are dedicated to delivering professional and reliable multi-omics solutions to support your scientific endeavors.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







