Spatial Proteomics Service
Spatial proteomics is an advanced branch of proteomics that focuses on mapping the precise spatial distribution of proteins within cells and understanding their interactions with other molecules in their native cellular context. At MtoZ Biolabs, we leverage our extensive expertise and state-of-the-art instrumentation to offer Spatial Proteomics Services, ensuring comprehensive support for your research needs.
Proteins perform a vast array of functions that are inherently tied to their precise locations within the cellular architecture. The subcellular localization of a protein determines its interaction partners, access to substrates, and participation in specific biochemical pathways, thereby directly influencing its biological role. For instance, transcription factors such as p53 and NF-κB are localized in the nucleus, where they regulate gene expression and orchestrate DNA repair mechanisms in response to cellular stress. Spatial proteomics allows for the detailed analysis of these proteins' localization in the nucleus, helping to uncover how their movement and interactions impact cellular responses. Enzymes of the oxidative phosphorylation pathway, including cytochrome c oxidase and ATP synthase, are situated in the mitochondria, where they are essential for ATP production and the regulation of apoptosis. Spatial Proteomics Service helps researchers track these enzymes within the mitochondria, shedding light on how their positioning affects energy production and cellular signaling.
Mislocalization of proteins, such as the aberrant cytoplasmic accumulation of tau in neurodegenerative diseases or the misplacement of insulin receptors in metabolic disorders, can disrupt cellular homeostasis and contribute to the pathogenesis of various diseases, highlighting the importance of understanding protein distribution within cells and the ability to capture protein subcellular dynamics. Spatial proteomics provides advanced methodologies to precisely map and quantify protein localization dynamics at subcellular resolution, enabling a deeper understanding of protein function, cellular compartmentalization, and the molecular mechanisms that govern both physiological processes and disease pathogenesis.
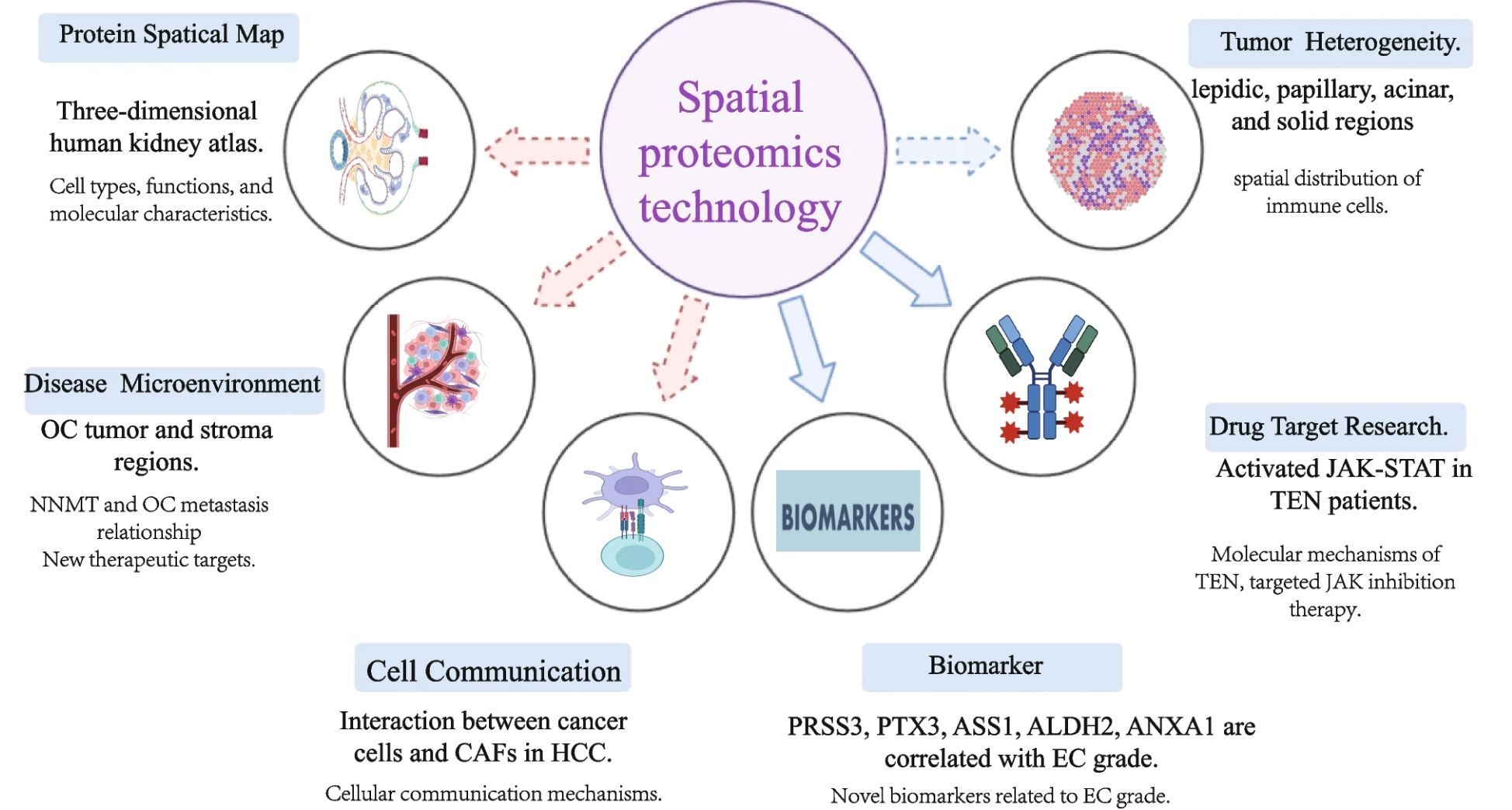
Figure 1. Examples of Related Findings and Troubleshooting in Spatial Proteomics
Services at MtoZ Biolabs
Spatial proteomics employs both mass spectrometry-based and imaging-based techniques to precisely map and analyze the distribution of proteins within cells and tissues. Mass Spectrometry-Based Methods, such as Imaging Mass Spectrometry (IMS), provide high-resolution quantitative data on protein localization and abundance without the need for labeling. Imaging-Based Methods, including Multiplexed Imaging Platforms and Super-Resolution Fluorescence Microscopy, enable the visualization of multiple proteins simultaneously, offering detailed insights into protein interactions and cellular architecture. These complementary approaches allow researchers to gain a comprehensive understanding of protein spatial dynamics, essential for unraveling complex biological processes and disease mechanisms. Leveraging these advanced methodologies and expert team, MtoZ Biolabs provides comprehensive Spatial Proteomics Services.
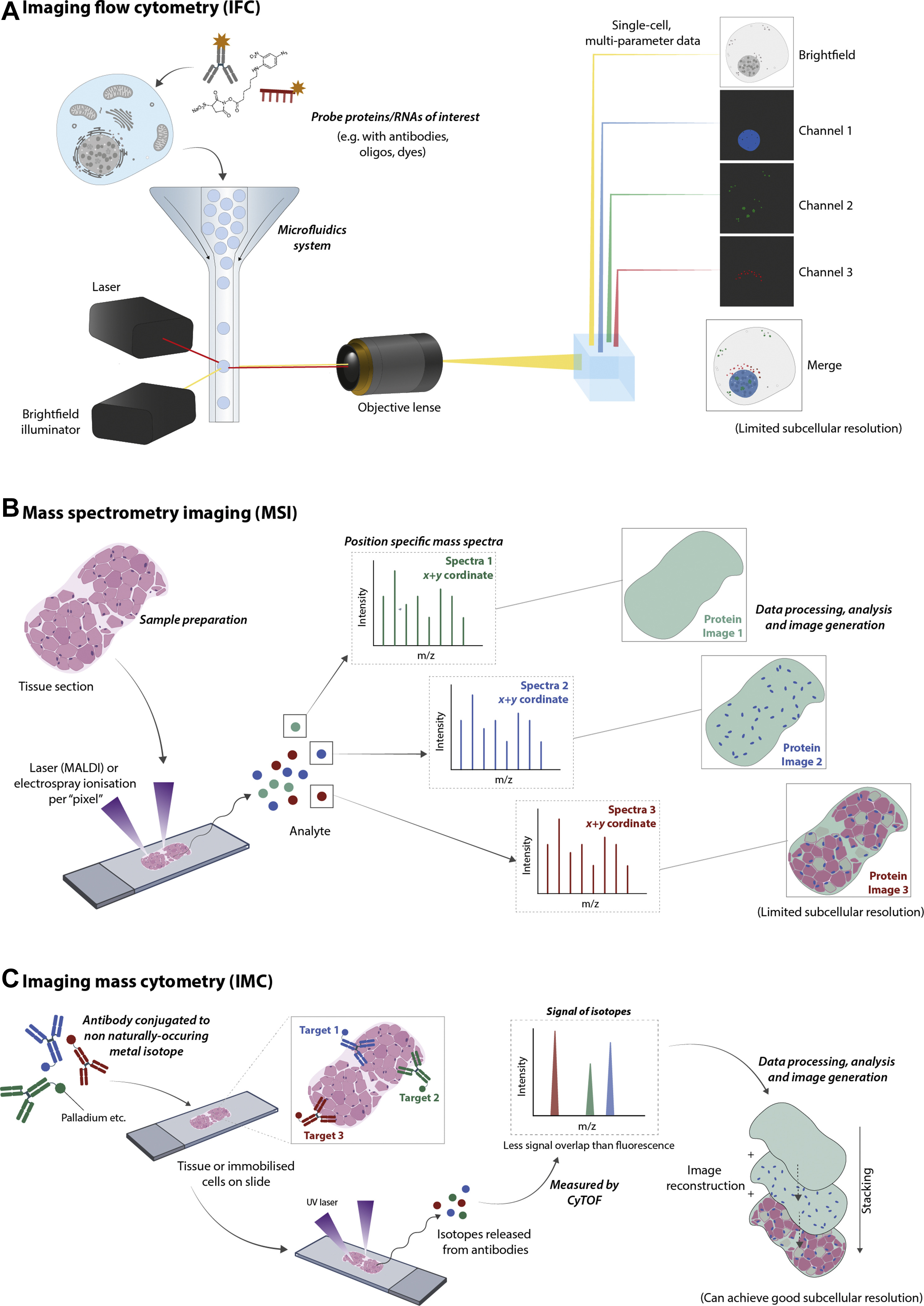
Christopher, J. A. et al. Mol Cell Proteomics. 2022.
Figure 1. Overview of the Major Spatial Proteomics Methods
Why Choose MtoZ Biolabs?
1. Advanced Analysis Platform: MtoZ Biolabs established an advanced Spatial Proteomics Service platform, guaranteeing reliable, fast, and highly accurate analysis service.
2. One-Time-Charge: Our pricing is transparent, no hidden fees or additional costs.
3. High-Data-Quality: Deep data coverage with strict data quality control. AI-powered bioinformatics platform integrates all spatial proteomics data, providing clients with a comprehensive data report.
4. Customized Research Solutions: Receive tailored services that address your unique research questions and experimental requirements.
5. Efficient Turnaround: Accelerate your research timeline with our prompt and efficient service delivery.
Sample Submission Suggestions
1. Sample Types: We accommodate both fresh-frozen (FF) and formalin-fixed paraffin-embedded (FFPE) tissue sections. Ensure that samples are properly preserved to maintain protein integrity and spatial distribution.
2. It is advised to prepare more than 3 biological replicates.
*Note: Please contact us if you have any special requirements or need assistance with your sample preparation.
Applications
1. Cancer Research: Map tumor microenvironment proteins to identify novel biomarkers and therapeutic targets.
2. Neuroscience: Analyze protein localization in neural tissues to understand brain function and neurological disorders.
3. Immunology: Profile immune cell protein interactions to elucidate mechanisms of immune response.
4. Drug Development: Identify and validate therapeutic targets through precise spatial protein analysis in disease models.
5. Developmental Biology: Investigate protein dynamics during tissue differentiation and embryonic development.
6. Pathology: Enhance diagnostic accuracy by spatially profiling proteins in diseased versus healthy tissues.
Case Study
1. Exploring Subcellular Protein Dynamics in Hepatic Steatosis Using MS-Based Comparative Spatial Proteomics
Hepatic steatosis, a hallmark of non-alcoholic fatty liver disease (NAFLD), involves significant alterations in subcellular protein localization, impacting hepatocyte function. To investigate these changes, a MS-based spatial proteomics study was conducted using mice fed either a normal diet or a high-fat diet to induce hepatic steatosis. Protein correlation profiling maps were generated to reveal striking subcellular translocations in the liver proteome. Key findings included the relocalization of numerous contact-site proteins to lipid droplets (LDs), highlighting the critical role of LDs in disease progression. Additionally, Golgi apparatus adsorption onto LDs was observed, leading to secretion defects—a potential molecular mechanism underlying reduced hepatocyte functionality in NAFLD. These spatial alterations shed light on how lipid accumulation disrupts cellular architecture and protein networks. This case demonstrates how MS-based spatial proteomics service provides detailed insights into protein dynamics, illuminating disease mechanisms and guiding therapeutic development.

Lundberg, E. et al. Nat Rev Mol Cell Biol. 2019.
Figure 3. Understanding the Cellular Consequences of Hepatic Steatosis
2. Imaging-Based Spatial Proteomics to Study Spatiotemporal Organization of Protein During Mitosis
Understanding the spatiotemporal organization of proteins during mitosis is crucial for unraveling the dynamics of cellular division. A recent study integrated 4D imaging with quantitative protein concentration measurements to investigate the behavior of 28 GFP-tagged human proteins in HeLa cells during mitosis. This imaging-based spatial proteomics approach generated a comprehensive model of protein reorganization across different mitotic phases, enabling detailed analysis of dynamic protein interactions. During cytokinesis, the study mapped protein colocalizations using a network graph, where nodes represented proteins and edges indicated dynamic localization clusters. Computational image analysis assigned edge colors to distinct localization clusters, providing a visual framework for understanding protein interplay. This approach revealed critical dynamic interactions, offering insights into how protein spatial organization governs key mitotic events. This case highlights the power of imaging-based spatial proteomics service to capture both spatial and temporal protein dynamics with exceptional precision.
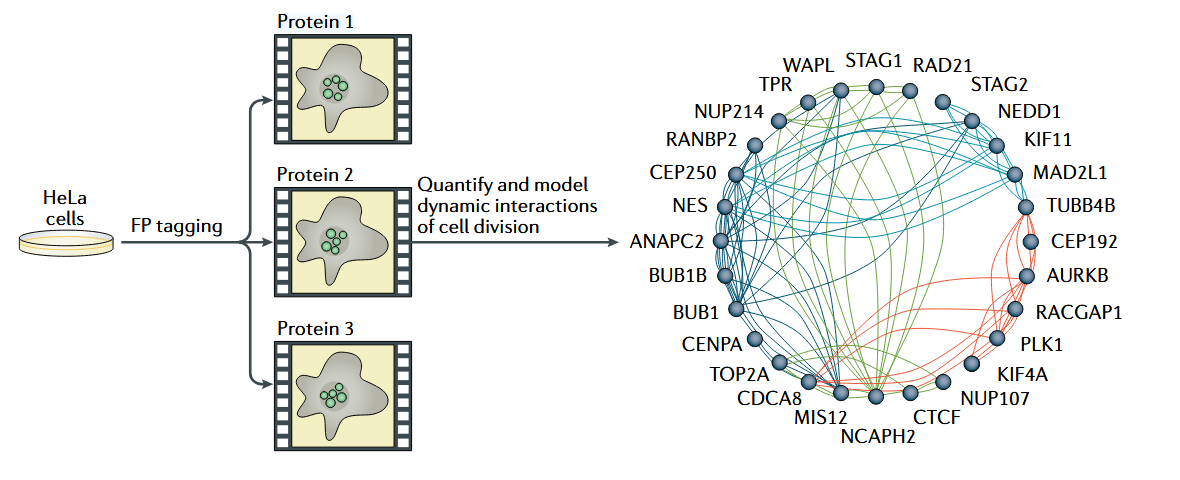
Lundberg, E. et al. Nat Rev Mol Cell Biol. 2019.
Figure 4. Spatiotemporal Organization of Human Proteins During Mitosis
Deliverables
1. Comprehensive Experimental details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Free project evaluation, welcome to learn more details!
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







