What Is the Difference Between Spatial Proteomics and Spatial Transcriptomics?
-
Spatial proteomics reveals what cells are doing at specific locations.
-
Spatial transcriptomics reveals what cells are preparing to do at specific locations.
-
Directly measures functional proteins, providing results that more closely reflect biological phenotypes.
-
Independent of transcriptional activity, capable of capturing post-translational regulation and modifications.
-
Limited throughput, especially for antibody-dependent methods.
-
Lack of standardized protocols, leading to complex data processing.
-
High-throughput with wide coverage, suitable for constructing spatial cell atlases.
-
Highly compatible with single-cell RNA sequencing, enabling integrated analyses.
-
mRNA expression does not always correspond to protein levels, potentially leading to discrepancies.
-
Achieving both high spatial resolution and broad tissue coverage is challenging.
Spatial proteomics and spatial transcriptomics are two fundamental technologies in the field of spatial omics. Both aim to characterize the molecular distribution within cells and tissues in a spatially resolved manner, but they differ in the types of biomolecules analyzed and their technical strategies.
Definitions and Research Objects
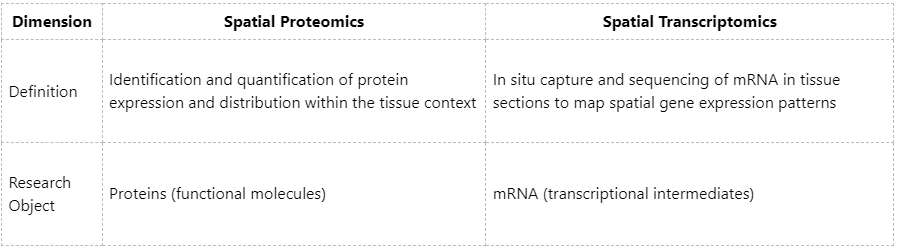
In summary:
Technical Principles and Methods
1. Spatial Proteomics
(1) Antibody-Based Methods
Examples: CODEX, MIBI, IMC (Imaging Mass Cytometry)
These approaches use metal-labeled antibodies to stain tissues and detect multiple protein signals through mass spectrometry imaging.
(2) Mass Spectrometry Imaging Methods
Examples: MALDI-MSI, timsTOF fleX
These methods do not require antibodies; proteins or peptides are directly detected on tissue sections for spatial mapping.
(3) Laser Microdissection Coupled with Mass Spectrometry
Specific tissue regions are isolated using laser microdissection, proteins are extracted, and quantitative analysis is performed using LC-MS/MS.
2. Spatial Transcriptomics
(1) In Situ Capture Methods (Mainstream)
Examples: 10x Genomics Visium, Slide-seq, Stereo-seq
Pre-spotted primers on slides capture mRNA in situ, which is then sequenced to assign spatial coordinates to transcripts.
(2) In Situ Hybridization Methods
Examples: MERFISH, seqFISH+
Fluorescent probes target specific mRNAs, and multiple rounds of imaging allow the reconstruction of their spatial distributions.
Resolution and Throughput

Explanation:
Spatial proteomics is constrained by the availability of antibodies or the detection capacity of mass spectrometry, resulting in relatively low throughput. Spatial transcriptomics can capture the entire transcriptome, although spatial resolution may require trade-offs.
Advantages and Limitations
1. Spatial Proteomics
(1) Advantages
(2) Limitations
2. Spatial Transcriptomics
(1) Advantages
(2) Limitations
Comparison of Practical Application Scenarios
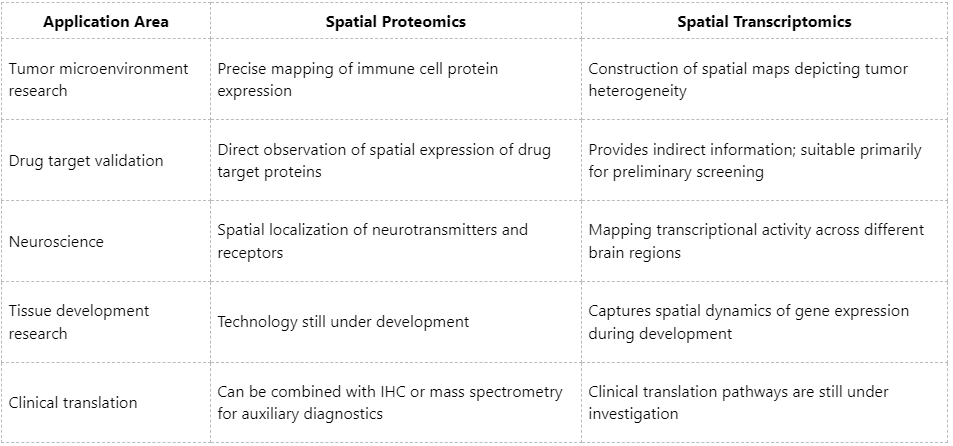
Although spatial proteomics and spatial transcriptomics each have distinct advantages and limitations, they are complementary rather than mutually exclusive. Increasingly, studies are integrating both approaches to develop spatial multi-omics platforms, enabling more comprehensive and precise characterization of tissue microenvironments. At MtoZ Biolabs, we provide end-to-end solutions covering spatial proteomics, spatial transcriptomics, and spatial metabolomics. Leveraging advanced imaging mass spectrometry and high-resolution single-cell platforms, we assist clients in achieving precise analysis from tissue to molecular level, and from spatial mapping to mechanistic understanding, with broad applications in oncology, immunology, neuroscience, and metabolism. Contact Biotech Paike for customized experimental design and data analysis services.
How to order?







