What Is the Difference between LFQ and iBAQ?
-
Does not require isotopic or chemical labeling, enabling streamlined experimental workflows.
-
Well suited for large-scale comparative analyses across multiple experimental conditions.
-
Strongly dependent on robust experimental design, data consistency, and statistical rigor.
-
Incorporates protein length and theoretical peptide numbers, improving abundance normalization across proteins.
-
Particularly suitable for proteome-wide expression profiling, computational model construction, and inference of cellular composition.
-
Conceptually closer to absolute quantification, while still relying on MS signal intensity as an indirect surrogate.
-
Deployment of high-resolution mass spectrometry platforms, including Orbitrap Fusion Lumos.
-
Support for both LFQ- and iBAQ-based quantitative workflows.
-
Comprehensive services encompassing data quality control, statistical evaluation, and biological interpretation.
- Across mechanistic studies, clinical translational research, and bioinformatics modeling, highly accurate and reproducible quantitative proteomics data can be generated.
In quantitative proteomics, LFQ (Label-Free Quantification) and iBAQ (Intensity-Based Absolute Quantification) represent two widely applied yet conceptually distinct quantification strategies. Although both approaches are derived from DDA (Data-Dependent Acquisition) mass spectrometry data, they differ markedly with respect to quantitative objectives, computational principles, and interpretation of results. A clear understanding of these differences is essential for selecting an appropriate quantification strategy tailored to specific research goals.
Basic Principles: LFQ vs. iBAQ
1. LFQ (Label-Free Quantification)
LFQ is a relative quantification approach designed to compare changes in the abundance of the same protein across multiple samples. The fundamental principle of LFQ involves normalizing and comparing MS1 signal intensities (typically peak areas) of peptides derived from identical proteins across different samples, thereby enabling inference of differential protein expression.
(1) Core Logic: Emphasizes relative abundance changes of the same protein across distinct biological samples.
(2) Output: Unitless, normalized abundance values for each protein across samples.
(3) Key Characteristics
2. iBAQ (Intensity-Based Absolute Quantification)
iBAQ is a quasi-absolute quantification strategy aimed at estimating relative abundance relationships among different proteins within a single sample. The method calculates protein abundance by summing the MS1 intensities of all identified peptides corresponding to a protein and normalizing this value by the theoretical number of peptides that the protein is expected to generate upon digestion.
(1) Core Logic: Reflects relative molar abundance relationships among proteins within the same biological sample.
(2) Output: Relative molar protein abundance, enabling quantitative comparisons across proteins.
(3) Key Characteristics
Overview of Core Differences
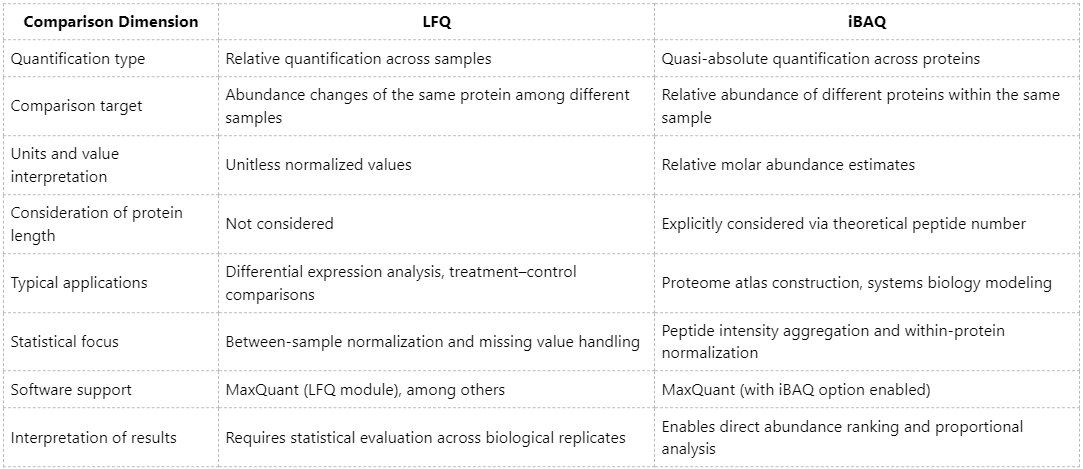
LFQ vs. iBAQ: Which Method Should Be Chosen?
When the primary objective is to identify differentially expressed proteins between experimental conditions, LFQ is generally preferred due to its robustness in comparative statistical analysis. In contrast, when the research focus lies in characterizing the abundance composition within a single sample-such as functional category proportions or the relative contribution of structural versus enzymatic proteins, iBAQ offers distinct advantages. Rather than being interchangeable, LFQ and iBAQ address complementary biological questions. Selection of the appropriate method should therefore be guided by experimental objectives, and in some cases, the combined application of both approaches may yield a more comprehensive quantitative perspective.
Quantitative Strategy Support at MtoZ Biolabs
At MtoZ Biolabs, quantitative proteomics strategies are tailored to specific research needs:
LFQ primarily captures abundance changes between experimental groups, whereas iBAQ characterizes the overall structure of protein abundance within a sample. These two approaches differ substantially with respect to quantitative focus, interpretation of outputs, and applicable research contexts. A thorough understanding of these distinctions facilitates more rigorous experimental design and more accurate interpretation of quantitative proteomics data. For researchers with related project requirements or technical inquiries, MtoZ Biolabs offers professional support and dedicated technical consultation.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







