Label-Free Quantitative Proteomics Service | Quantitative Phosphoproteomics
- Kinase-Substrate Interaction Studies: Identify phosphorylation targets and uncover kinase activity dynamics in signaling pathways.
- Signal Transduction Research: Quantify phosphorylation changes to investigate regulatory mechanisms in cellular processes.
- Disease Biomarker Discovery: Detect differentially phosphorylated proteins linked to cancer, neurodegeneration, and metabolic disorders.
- Drug Response and Mechanism Studies: Evaluate phosphorylation changes induced by kinase inhibitors and other therapeutic agents.
- Pathway and Network Analysis: Map phosphorylation-driven signaling cascades to understand cellular regulation.
Label-Free Quantitative Proteomics (LFQ) is a powerful mass spectrometry-based approach for quantifying protein abundance across multiple biological conditions without the need for isotope labeling. Unlike label-based methods such as SILAC and TMT, label-free quantitative proteomics determines protein and peptide levels by measuring signal intensity or spectral counts, providing a cost-effective, scalable, and highly flexible solution. Label-free quantitative proteomics is particularly suited for large-scale studies, time-course experiments, and clinical research, where labeling is impractical. Advances in high-resolution LC-MS/MS instrumentation and bioinformatics-driven normalization techniques have significantly improved the accuracy and reproducibility of label-free quantitative proteomics, making it a preferred choice for global protein quantification.
Phosphorylation is a dynamic and reversible post-translational modification that regulates protein activity, stability, and interactions, playing a key role in cellular signaling and disease processes. Understanding phosphorylation dynamics requires precise quantification of phosphopeptides across biological conditions. Traditional labeling methods, such as SILAC and TMT, offer accurate quantification but are costly, labor-intensive, and unsuitable for certain sample types. In contrast, label-free quantitative proteomics provides a flexible and cost-effective alternative, allowing large-scale phosphorylation studies without the constraints of isotope labeling.
Service at MtoZ Biolabs
Precise quantification of phosphorylated peptides is crucial for understanding kinase activity, signaling dynamics, and disease mechanisms. Phosphorylation events are often low in abundance and highly transient, requiring specialized enrichment techniques such as IMAC and TiO₂ chromatography before LFQ analysis. MtoZ Biolabs offers a comprehensive Label-Free Quantitative Proteomics Service that integrates optimized phosphopeptide enrichment with cutting-edge LC-MS/MS and advanced bioinformatics to deliver high-confidence phosphorylation quantification. Whether investigating kinase-substrate interactions, cellular signaling pathways, or drug responses, the Label-Free Quantitative Proteomics Service provides the sensitivity and throughput necessary for comprehensive phosphoproteomic research.
Analysis Workflow
1. Sample Preparation & Protein Extraction: Total proteins are extracted while preserving phosphorylation states. Protein concentration and quality are assessed to ensure sample integrity.
2. Protein Digestion & Phosphopeptide Enrichment: Proteins are first digested into peptides, followed by phosphopeptide enrichment using IMAC, TiO₂, immunoprecipitation, or other affinity-based methods.
3. Fractionation & LC-MS/MS Analysis: Enriched phosphopeptides are fractionated, and high-resolution LC-MS/MS is then used to identify and quantify phosphorylation events.
4. Label-Free Quantification & Data Processing: Phosphopeptides are quantified based on intensity or spectral counts. Advanced normalization ensures reproducibility and minimizes batch effects.
5. Bioinformatics & Statistical Analysis: Identified phosphosites are mapped to kinases, pathways, and cellular functions. Statistical models detect significant phosphorylation changes, providing insights into signaling dynamics and disease mechanisms.
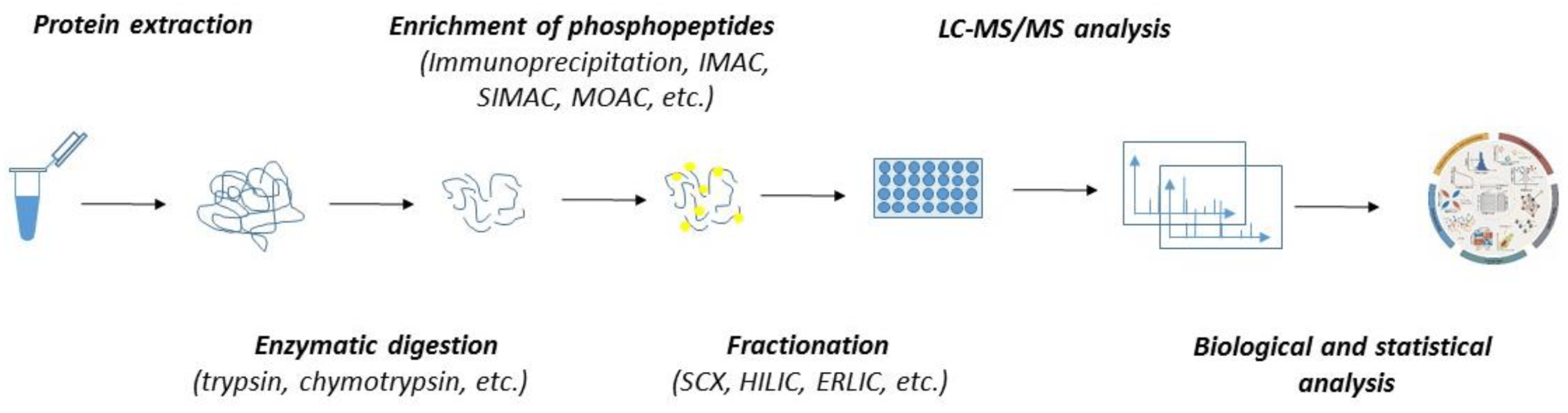
Figure 1. Workflow of Label-Free Quantitative Proteomics Service
Service Advantages
✔️High-Throughput Phosphorylation Analysis: Enables large-scale quantification of phosphosites with deep coverage, detecting thousands of phosphorylation events.
✔️Cost-Effective and Flexible Quantification: Unlike isotope-labeling methods, Label-Free Quantitative Proteomics Service eliminates the need for expensive reagents and enables large-scale phosphorylation studies across diverse sample types.
✔️Optimized Phosphopeptide Enrichment: Utilizes IMAC, TiO₂, and antibody-based methods to maximize phosphopeptide recovery and specificity.
✔️Customizable Workflows: Tailored experimental designs and expert guidance in our Label-Free Quantitative Proteomics Service support diverse research needs, from basic signaling studies to biomarker discovery and drug development.
Applications
Our Label-Free Quantitative Proteomics Service provides precise phosphorylation quantification, enabling researchers to explore signaling dynamics and disease mechanisms across various biological contexts.
Case Study
Investigating Tyrosine Phosphorylation in Sperm Capacitation Using Label-Free Quantitative Proteomics
Sperm capacitation is essential for fertilization and involves increased tyrosine phosphorylation, yet its regulatory mechanisms remain unclear. Using Label-Free Quantitative Proteomics Service, researchers mapped phosphorylation changes in human sperm during capacitation via high-resolution LC-MS/MS. This approach identified 231 phosphosites with increased levels, with motif analysis revealing IGF1R/insulin receptor kinases as key regulators. Inhibition of IGF1R with GSK1904529A and NVP-AEW541 suppressed tyrosine phosphorylation and sperm hyperactivated motility, while IGF1 stimulation restored both functions. These findings highlight the IGF1R-mediated phosphorylation pathway as a crucial regulator of sperm capacitation. This study suggests IGF1R as a potential therapeutic target for improving sperm function in infertility treatments and demonstrates the power of Label-Free Quantitative Proteomics Service in reproductive biology.
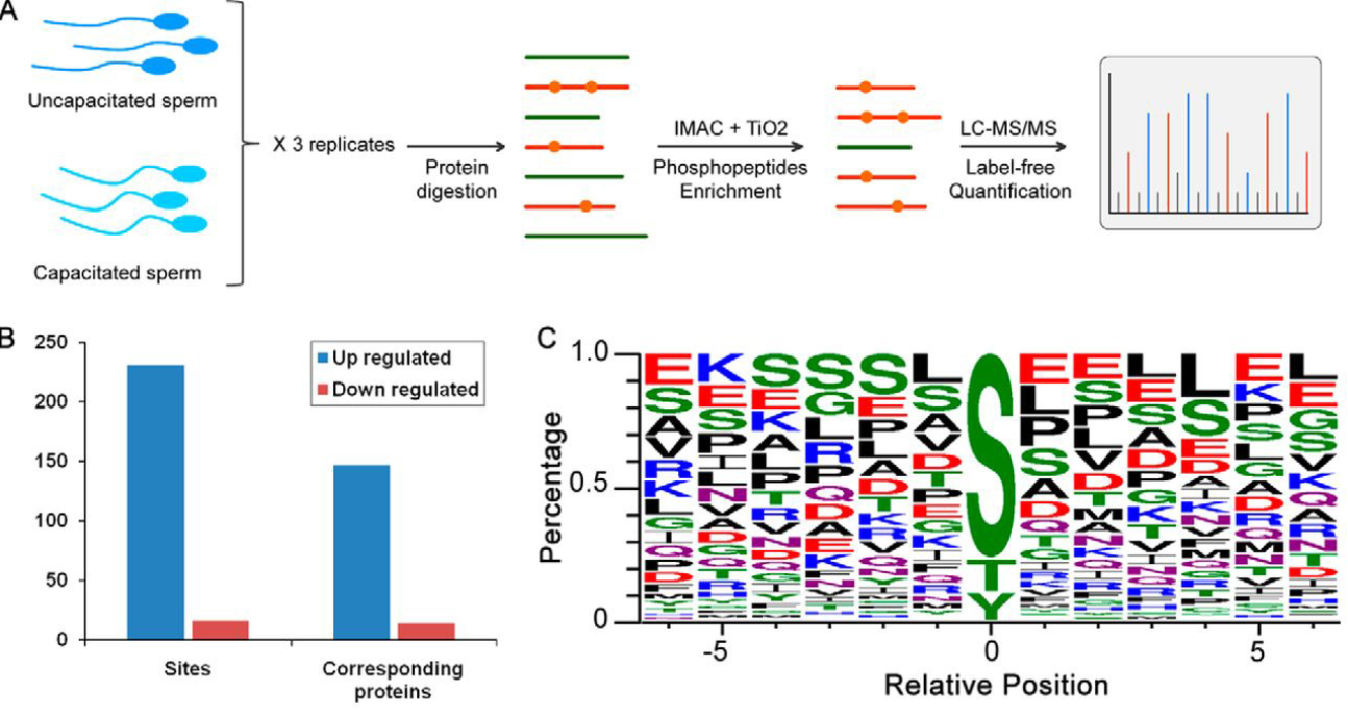
Figure 2. Label-Free Quantitative Phosphoproteomics Profiling of Human Sperm Capacitation
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment
4. Data Analysis, Preprocessing, and Estimation
5. Bioinformatics Analysis
6. Raw Data Files
With an unwavering commitment to scientific rigor, MtoZ Biolabs' Label-Free Quantitative Proteomics Service leverages advanced technologies, expert guidance, and robust data analytics to provide deep insights into complex protein signaling pathways. Feel free to reach out for consultation on your specific needs.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Label-Free Quantitative Proteomics Service, MS Based
How to order?







