What Is Non‑Degradative Ubiquitination?
-
Altering protein conformation to activate or inhibit function: For example, K63-linked ubiquitination of IKKγ promotes activation of the IKK complex, thereby initiating NF-κB signaling.
-
Regulating subcellular localization: Ubiquitin can act as a molecular address label, directing proteins to specific compartments such as the nucleus or lysosomes.
-
Mediating protein–protein interactions: Certain ubiquitin chain conformations create docking platforms for specialized recognition domains, including UBA and NZF motifs.
-
Dynamic reversibility: Ubiquitination can be rapidly reversed by deubiquitinating enzymes (DUBs), conferring high plasticity to the regulation of signaling pathways.
-
Ubiquitin site mapping (UbiSite/diGly analysis): Identifies modification sites by detecting the diglycine remnant derived from ubiquitin.
-
Ubiquitin chain type characterization: Utilizes chain-specific antibodies or enzymatic cleavage approaches to discriminate between K63-, K11-, and M1-linked chains.
-
Dynamic quantification: Measures changes in ubiquitination levels under varying stimulation conditions to uncover underlying regulatory mechanisms.
-
Comprehensive ubiquitination profiling: Coverage of all chain types with precise site identification.
-
Quantitative comparative analyses: Supporting labeling strategies such as SILAC and TMT to reveal dynamic changes in ubiquitination.
-
Ubiquitin substrate screening: Integration of immunoenrichment and mass spectrometry to identify candidate substrate proteins.
-
End-to-end technical and data analysis support: Encompassing experimental design through to bioinformatics interpretation.
In eukaryotic cells, ubiquitination represents a pivotal post-translational modification whereby the small protein ubiquitin is covalently conjugated to substrate proteins, thereby modulating their stability, activity, subcellular localization, and interaction networks. While the canonical and most extensively characterized role of ubiquitination is to direct proteins toward proteasomal degradation, typically via K48-linked polyubiquitin chains in the 26S proteasome pathway, accumulating evidence has established that non-degradative ubiquitination exerts more intricate and fine-tuned regulatory effects on diverse cellular processes.
Non-degradative ubiquitination is defined as the covalent attachment of ubiquitin to a substrate protein without promoting its degradation. Instead, it modulates protein function through mechanisms such as altering conformation, influencing spatial distribution, or recruiting specific interaction partners. This form of modification participates in a wide range of cellular events, including signal transduction, DNA damage repair, autophagy, and regulation of inflammation. Non-degradative ubiquitination is characterized by high reversibility, dynamic turnover, structural diversity in ubiquitin chain topology, and functional specificity, thereby constituting an integral component of the complex intracellular regulatory network.
Definition and Classification of Non-Degradative Ubiquitination
Non-degradative ubiquitination refers to ubiquitin conjugation events that do not target the substrate protein for degradation, but instead regulate its biological functions through alternative pathways. Based on the linkage pattern of ubiquitin chains and the number of ubiquitin moieties involved, it can be classified into the following types:
1. Monoubiquitination
The covalent attachment of a single ubiquitin molecule to a specific lysine residue of the target protein. This modification plays crucial roles in regulating protein localization (e.g., nuclear import), endocytosis, transcriptional regulation, and DNA repair.
2. Multi-Monoubiquitination
The conjugation of multiple ubiquitin molecules to distinct lysine residues within the same substrate. This pattern is frequently observed in the regulation of membrane protein endocytosis, such as in the downregulation of the epidermal growth factor receptor (EGFR).
3. Non-K48 Polyubiquitin Chain Modifications
Ubiquitin contains seven lysine residues (K6, K11, K27, K29, K33, K63, K48) and an N-terminal methionine (M1), enabling the assembly of polyubiquitin chains with diverse linkages:
(1) K63-linked chains: The most prevalent non-degradative linkage type, broadly involved in signaling pathways (e.g., NF-κB activation), DNA damage response, and autophagy.
(2) K11/K27/K29-linked chains: Exhibit distinct regulatory roles in cell cycle progression, developmental processes, and immune signaling.
(3) Linear (M1-linked) chains: Synthesized by the linear ubiquitin chain assembly complex (LUBAC) and critical for inflammatory responses and apoptosis regulation.
Molecular Mechanisms and Signaling Functions
Non-degradative ubiquitination modulates cellular functions through several distinct mechanisms:
Biological and Pathological Significance
Non-degradative ubiquitination is essential for numerous physiological processes, and dysregulation of its pathways can contribute to the pathogenesis of a variety of severe diseases, including those involving aberrant signaling, defective DNA repair, impaired immune responses, and autophagy-related disorders.
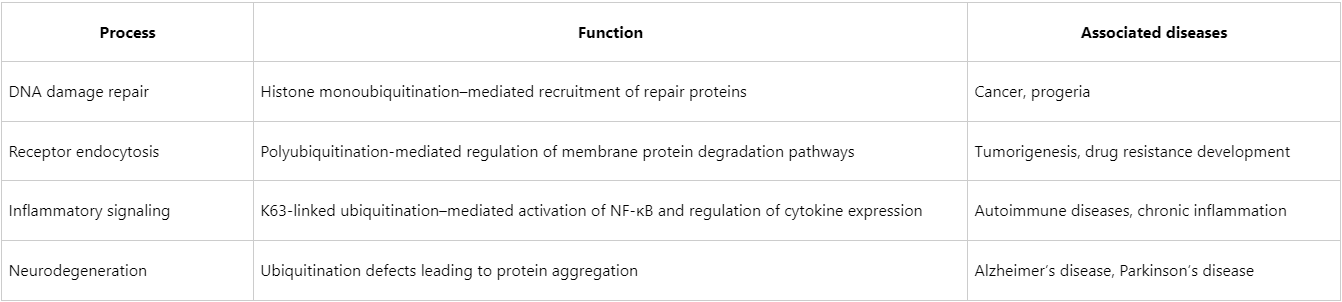
Research Strategies and the Role of Mass Spectrometry
Deciphering ubiquitination events, particularly the linkage sites and chain topologies of non-degradative modifications, remains a major challenge. High-resolution mass spectrometry (MS), especially LC-MS/MS-based platforms, has emerged as an indispensable analytical tool:
MtoZ Biolabs’s Solutions
In the field of ubiquitination research, particularly with respect to non-degradative pathways, MtoZ Biolabs offers the following specialized services:
By integrating state-of-the-art Orbitrap high-resolution MS platforms with standardized experimental workflows, we deliver high-sensitivity and highly reproducible ubiquitination analyses. This enables detailed investigation of ubiquitin’s regulatory roles in signaling networks and disease mechanisms. Non-degradative ubiquitination is not merely a post-translational modification but serves as a molecular code for intracellular information transfer. With the rapid advancement of high-throughput MS and proteomics technologies, elucidating the fine-scale regulation of this process is becoming a prominent frontier in life sciences research. MtoZ Biolabs remains committed to providing robust technical platforms and research support to facilitate significant breakthroughs in ubiquitination studies.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







