What Is Co-Immunoprecipitation (Co-IP)?
- Immunoprecipitation enrichment of the target complex
- SDS-PAGE separation or in-solution enzymatic digestion
- High-resolution mass spectrometric analysis (e.g., Orbitrap)
- Bioinformatic identification of interacting proteins
- Analysis of signaling pathways (e.g., identifying interacting partners of EGFR or MAPK)
- Investigation of transcriptional regulation (e.g., interactions between transcription factors and co-regulators)
- Elucidation of disease mechanisms (e.g., tumor-associated protein networks)
- Drug-target discovery and validation
In routine laboratory research focused on protein function and signal transduction pathways, co-immunoprecipitation (Co-IP) is a fundamental technique encountered by nearly all life-science researchers. It is widely regarded as one of the gold-standard methods for validating protein–protein interactions and serves as an essential bridge linking immunology with modern proteomics and mass spectrometry technologies.
Fundamental Principle of Co-Immunoprecipitation
Co-immunoprecipitation is an immunoaffinity purification technique based on the specific interaction between an antibody and its target (bait) protein. The key objective is to isolate a target protein together with its associated binding partners, thereby capturing the native protein complex indirectly. Compared with other interaction-detection methods such as yeast two-hybrid, FRET, or BiFC, Co-IP offers the advantage of being performed under near-physiological conditions, which helps preserve native protein conformations and interaction states.
Experimental Workflow of Co-Immunoprecipitation
1. Cell Lysis
Cells are lysed under mild, non-denaturing conditions (e.g., using non-ionic detergents such as NP-40 or Triton X-100) to preserve native protein complexes.
2. Antibody Binding
A specific antibody is added to the lysate to bind the bait protein and form an antibody–antigen complex.
3. Capture with Protein A/G Agarose Beads
Agarose beads coated with Protein A or Protein G are used to immobilize and capture the antibody complex.
4. Washing and Elution
After removing non-specific contaminants, the bound protein complex is eluted under denaturing conditions (for example, with SDS-PAGE loading buffer).
5. Detection of Interacting Proteins
Co-precipitated proteins are typically detected by Western blotting. Alternatively, mass spectrometry can be applied to identify previously uncharacterized interaction partners.
Comparison with Other Protein-Interaction Techniques
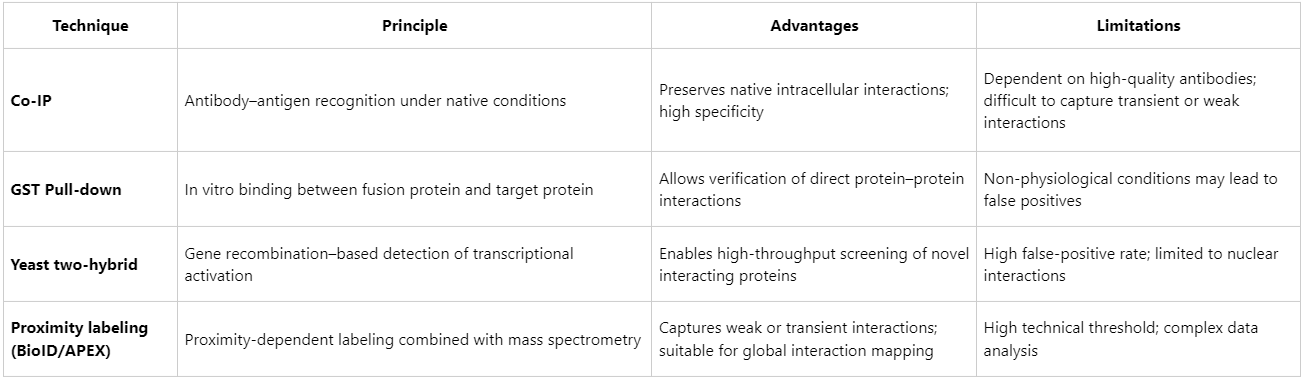
In practical applications, Co-IP remains one of the benchmark methods for confirming candidate protein interactions. It is particularly well-suited for large-scale interactome analyses when integrated with mass spectrometry.
Co-Immunoprecipitation Coupled with Mass Spectrometry: Exploring Protein Interaction Networks
With the rapid advancement of mass spectrometry technologies, Co-IP combined with mass spectrometry (Co-IP-MS) has become an effective strategy for characterizing the composition and dynamics of protein complexes. The typical workflow includes:
For instance, MtoZ Biolabs' advanced platforms equipped with high-sensitivity Orbitrap Exploris 480 instruments and optimized immuno-enrichment protocols, supporting diverse tag systems such as FLAG, HA, Myc, or GFP, enable high-throughput mapping of protein interaction networks.
Critical Considerations in Co-Immunoprecipitation Experiments
Although Co-IP is widely used, the reliability of results depends heavily on experimental design and technical parameters. Key factors include:
1. Antibody Quality
High-specificity and high-affinity antibodies are essential for successful immunoprecipitation. Commercially validated antibodies or pre-coated magnetic beads are recommended to enhance efficiency.
2. Lysis Conditions
The composition of the lysis buffer, particularly salt concentration and detergent type, should be optimized to maintain protein–protein interactions without excessive disruption.
3. Control of Non-Specific Binding
Including IgG and bead-only controls helps eliminate background noise. Sequential re-immunoprecipitation (Re-IP) can further improve specificity.
4. Endogenous versus Overexpressed Proteins
Although overexpression systems often yield stronger signals, they may introduce artificial interactions. Therefore, key findings should be validated using endogenous proteins whenever possible.
Applications of Co-Immunoprecipitation
Co-IP has broad applications across multiple areas of life science research, including but not limited to:
For researchers seeking comprehensive interactome analysis, Co-IP integrated with mass spectrometry and bioinformatic workflows provides an efficient, systematic approach for uncovering molecular mechanisms and accelerating scientific discovery.
As a powerful method for studying protein–protein interactions, co-immunoprecipitation (Co-IP) remains indispensable in molecular biology research due to its simplicity, specificity, and reproducibility. When coupled with high-resolution mass spectrometry, Co-IP greatly extends the depth and scope of protein-interaction studies, serving as a vital link between molecular mechanism research and systems-level biological exploration. If you require experimental support for protein interaction validation or high-throughput interactome analysis, MtoZ Biolabs offers efficient and reliable technical solutions to advance your research.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







