Variable Temperature Circular Dichroism (VT-CD) Analysis Service
- Unfolding cooperativity (single- vs. multi-phase transitions),
- Reversibility of thermal denaturation,
- Buffer and additive effects on structural stability, and
- Ligand-induced stabilization or destabilization.
Circular dichroism (CD) spectroscopy is widely recognized as a sensitive method for assessing the secondary structure of chiral biomolecules. When integrated with a temperature-controlled environment, variable temperature circular dichroism (VT-CD) enables real-time monitoring of conformational changes across a defined temperature gradient, offering insight into the cooperativity and reversibility of structural transitions. This approach is particularly valuable for characterizing protein unfolding/refolding transitions, determining melting temperatures (Tm), and evaluating the thermal stability of nucleic acid structures and protein–ligand complexes.
Technical Principles
VT-CD operates on the fundamental principle of circular dichroism, which measures the differential absorption of left- and right-handed circularly polarized light by optically active molecules. The resulting spectrum reflects the chiroptical properties associated with specific secondary structural elements—such as α-helices (208 nm, 222 nm), β-sheets (~215 nm), and random coils (~198 nm).
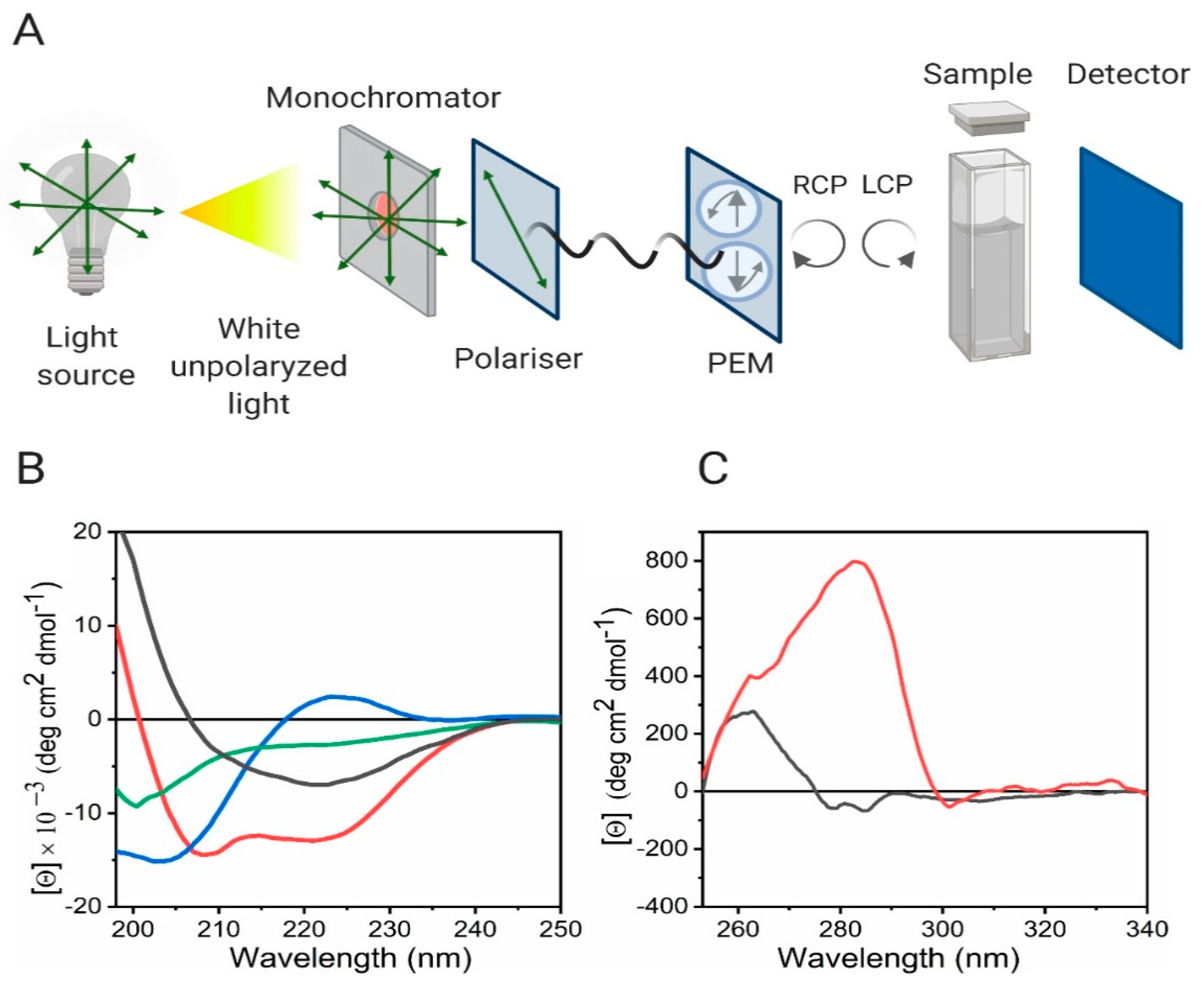
Pignataro, M.F. et al. Molecules. 2020.
Figure 1. Circular Dichroism as a tool for the study of protein secondary and tertiary structure
By recording CD spectra at incremental temperature points, typically ranging from 4°C to 95°C, VT-CD tracks structural transitions that occur as thermal energy disrupts non-covalent interactions stabilizing the native conformation. The change in ellipticity at characteristic wavelengths is plotted as a function of temperature to generate melting curves, from which key thermodynamic parameters—including Tm (midpoint of thermal transition) and Δθ (magnitude of conformational shift)—can be extracted.
This technique allows detailed analysis of:
Services at MtoZ Biolabs
At MtoZ Biolabs, we provide advanced VT-CD analysis services powered by high-precision CD instrumentation coupled with programmable temperature control. Our service enables researchers to analyze secondary structure transitions and determine thermal denaturation parameters such as melting temperature (Tm) to support a wide range of structural biology and biopharmaceutical applications:
1. Thermal unfolding and refolding studies for recombinant proteins and peptides
2. Secondary structure estimation across variable temperatures
3. Determination of melting temperature (Tm) and thermodynamic transition points
4. Stability analysis of native proteins, antibody domains, DNA/RNA structures
5. Comparative structural analysis under different formulation or buffer conditions
6. Ligand- or cofactor-induced stability assessment
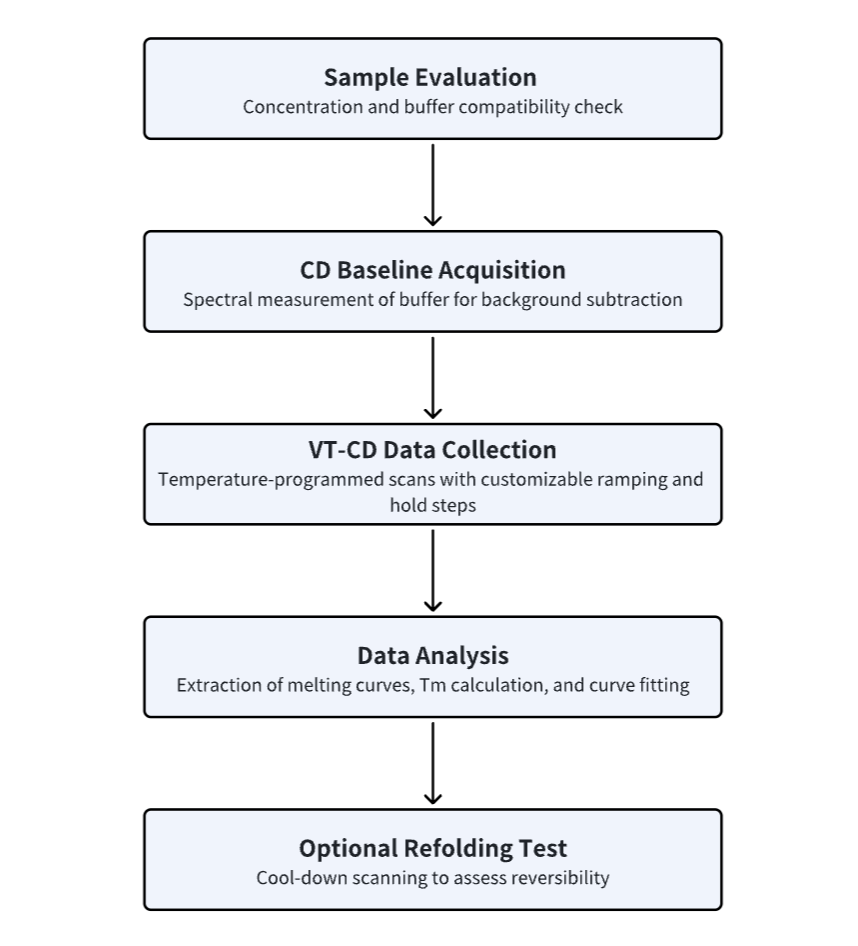
Analysis Workflow
Service Advantages
✅Precise temperature control (Peltier system, 0.1°C resolution)
✅Low sample consumption (typically <300 μL)
✅Real-time spectral tracking for unfolding/refolding
✅Suitable for proteins, nucleic acids, and protein–ligand complexes
✅Customizable scan parameters and temperature ranges
✅Detailed data interpretation and reporting
Sample Submission Suggestions
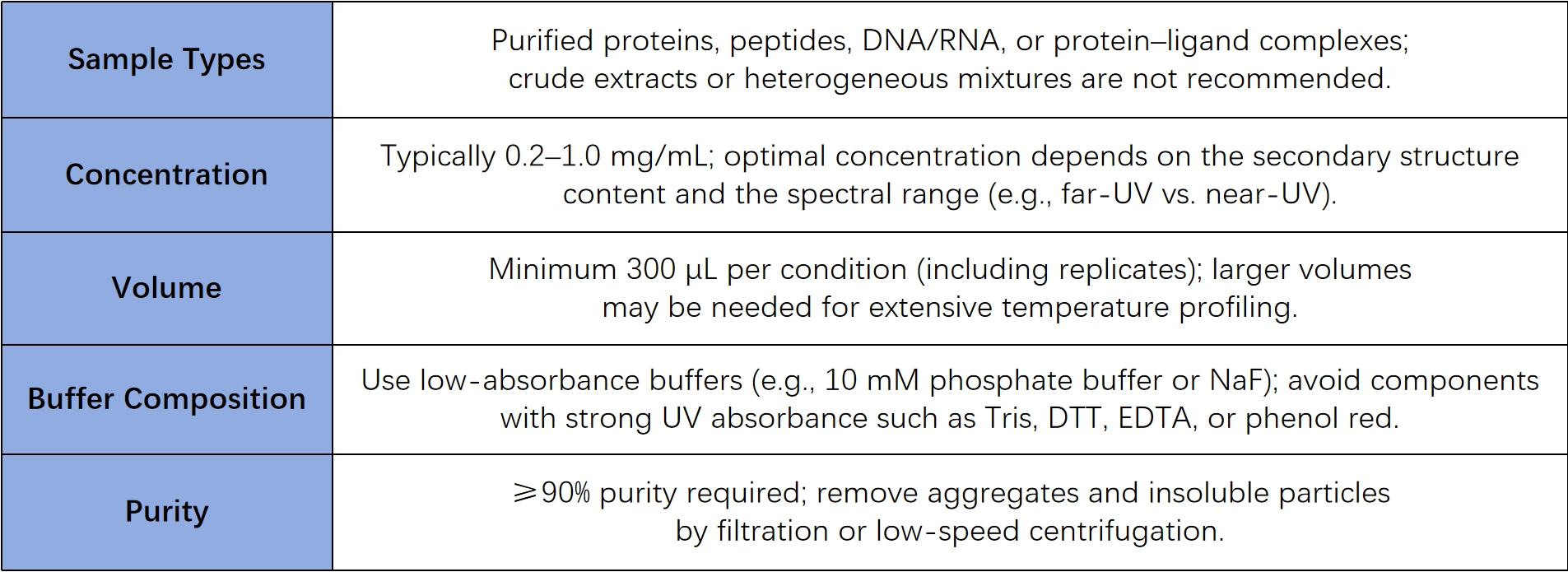
*Shipping and Storage: Store at –80°C and ship on dry ice. Avoid repeated freeze-thaw cycles. Cold-chain shipping (2–8°C) is acceptable if sample stability is ensured.
Applications
✅Protein Folding/Unfolding Analysis
Evaluate protein thermal stability (Tm) by monitoring temperature-dependent changes in α-helical signals (208 nm and 222 nm) or β-sheet signals (~215 nm).
✅Nucleic Acid Secondary Structure Melting Analysis
Investigate thermal melting behavior of structures such as G-quadruplexes and i-motifs.
✅Ligand-Induced Stability Assessment
Determine whether ligand binding alters protein conformation or enhances structural stability.
✅Thermal Refolding Experiments
Assess the reversibility of protein unfolding by monitoring structural recovery upon cooling after heat denaturation.
Related Service
Circular Dichroism Spectrum Analysis Service
Protein Structure Characterization Service
X-Ray Crystallography Protein Structure Determination Service
How to order?







