TMT vs Label-Free Quantitative Proteomics
-
High throughput: Enables simultaneous analysis of up to 18 samples, ideal for large cohort studies.
-
High precision: Batch effects are minimized through sample pooling, ensuring data consistency.
-
Enhanced statistical power: Uniform sample backgrounds facilitate detection of subtle expression changes.
-
High cost: Reagent expenses are significant, particularly for large-scale studies.
-
Ratio compression: Quantitative accuracy may be compromised, especially for low-abundance proteins.
-
Technical complexity: Requires advanced instruments (e.g., Orbitrap Fusion Lumos) and MS3 optimization.
-
Cost-effective: No need for costly chemical tags, suitable for budget-conscious research.
-
Flexible experiment design: Samples can be analyzed in batches, accommodating long-term or dynamic studies.
-
Greater depth of coverage: Avoids losses due to labeling inefficiencies; may yield higher protein identification.
-
Reproducibility challenges: Susceptible to instrument drift and batch variability, requiring stringent QC.
-
Limited throughput: Increased sample numbers proportionally increase instrument time.
-
Quantification variability: Signal-to-noise ratio may be low for low-abundance peptides.
-
Precious sample studies (e.g., tumor tissues, plasma) with limited material requiring accurate quantification.
-
Integrative multi-omics research involving transcriptomics or metabolomics.
-
High-throughput clinical cohort analyses.
-
Drug target discovery or mechanism-of-action studies detecting subtle changes.
-
Exploratory or early-stage research.
-
High-throughput screening with ample sample availability.
-
Projects with limited funding requiring broad protein identification.
-
Time-course studies analyzing cellular responses at multiple time points.
-
Equipped with Thermo Orbitrap Fusion Lumos and Exploris 480 platforms, fully compatible with TMT and LFQ workflows.
-
Proprietary technologies for enriching low-abundance proteins, enhancing quantification sensitivity.
-
One-stop services including experimental design, sample QC, data mining, and bioinformatics analysis.
-
Dual-language reporting for all quantitative proteomics projects, facilitating international collaboration and publication.
Proteomics, as a pivotal tool for elucidating biological mechanisms, is transitioning from a discovery-driven approach to a phase of precise quantification. Among large-scale quantitative strategies, Tandem Mass Tag (TMT) labeling and Label-Free Quantification (LFQ) are the most widely adopted. Although both rely on high-resolution mass spectrometry platforms, they exhibit fundamental differences in experimental design, data quality, and suitable application scenarios.
What is TMT-Based Quantification?
TMT (Tandem Mass Tag) is a relative quantification technique in proteomics based on isobaric chemical labeling. It enables multiplexed sample analysis and quantitative comparison by covalently attaching isobaric tags with distinct reporter ions to peptides or proteins derived from different samples.
1. Brief Principle
Each TMT reagent consists of a reporter ion, a mass balancer, and a reactive group.
Following proteolytic digestion, peptides from each sample are labeled with distinct TMT tags and subsequently combined into a single mixture for LC-MS/MS analysis.
At the MS1 level, all labeled peptides exhibit identical masses due to the isobaric nature of the tags. During MS2 or MS3 fragmentation, specific reporter ions are released and used for relative quantification across samples.
2. Common TMT Reagents
TMT 6plex, 10plex, 11plex, and 16plex (with the latest being TMTpro 18plex) support multiplexed quantification of multiple samples in a single run, significantly enhancing throughput and consistency.
What is Label-Free Quantification (LFQ)?
Label-free quantification refers to methods that do not require chemical or metabolic labeling of samples. Instead, quantification is based on MS1 ion intensities or spectral counting from mass spectrometry data.
1. Brief Principle
Each sample is analyzed independently via LC-MS. The intensity of the MS signal is directly proportional to the abundance of peptides or proteins.
Quantitative comparisons are achieved by evaluating peak areas or spectral counts, followed by retention time alignment, peak detection, and normalization to ensure data comparability.
Core Differences Between TMT and Label-Free Proteomics
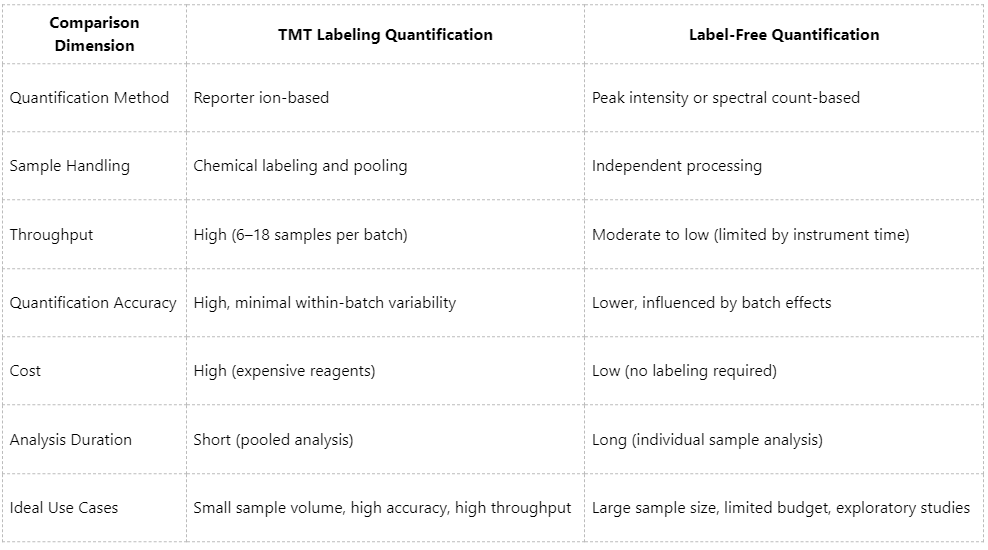
Advantages and Limitations: Choosing Between TMT and LFQ
1. TMT Advantages
2. TMT Limitations
3. LFQ Advantages
4. LFQ Limitations
Recommended Application Scenarios
1. Best Suited for TMT
2. Best Suited for LFQ
MtoZ Biolabs' Proteomics Solutions
Selecting between TMT and LFQ is often a decisive factor in the success of a proteomics project. MtoZ Biolabs offers tailored proteomics solutions to meet diverse research needs:
Both TMT and LFQ have their respective strengths. The choice of strategy should be guided by specific research objectives, budget constraints, and sample characteristics. TMT is optimal for high-throughput, high-precision comparisons, while LFQ offers flexibility and cost-efficiency for early-stage exploration. Selecting the appropriate quantification method can substantially enhance the efficiency and impact of proteomic investigations. For further consultation on experimental design or technical pathway selection, please feel free to contact MtoZ Biolabs, we are committed to supporting your scientific advancement with professional expertise and high-quality service.
How to order?







