Targeted Protein Quantification: Achieving Precision at the Peptide Level through PRM Technology
- Peptide selection and synthesis: Avoid peptides containing modification sites, sequence variants, or labile residues prone to degradation.
- Internal standard design and application: Internal standard peptides must exactly match target sequences, differing only by terminal isotope labels; they should be added prior to digestion to ensure complete process calibration.
- Chromatographic performance: Consistent peak shapes and retention times are fundamental to achieving linear and accurate quantification.
- Mass spectrometry parameters: Isolation window (1–2 Da), maximum injection time, and scan cycle must be optimized for the best analyte response.
- Background suppression and signal enhancement: Strategies such as multi-step elution, gradient optimization, and offline fractionation (e.g., SPE) effectively minimize co-elution interference.
- High-throughput target discovery: Integration of DIA data and in silico prediction to construct comprehensive peptide candidate libraries.
- SIL peptide customization and quality control: Custom synthesis of stable isotope–labeled peptides with rigorous validation of purity and quantification accuracy.
- Automated PRM workflow: A standardized pipeline implemented on the Skyline platform, covering target selection through parameter optimization.
- Compatibility with complex samples: Effective processing of plasma, tissue homogenates, and cerebrospinal fluid using advanced enrichment and cleanup techniques.
- Automated identification of optimal fragment ion combinations
- Dynamic adjustment of acquisition windows for improved sensitivity
- Cross-batch normalization for large-scale comparative studies
In proteomics research, achieving accurate and reproducible quantification of proteins of interest is essential for elucidating disease mechanisms, validating drug targets, and facilitating translational studies. However, due to the intrinsic complexity of proteins and the vast dynamic range of their expression levels, direct quantification at the protein level remains highly challenging. Consequently, peptide level quantification strategies have emerged as the predominant approach. Parallel Reaction Monitoring (PRM), a key mass spectrometry-based technique for targeted protein quantification, offers exceptional selectivity and sensitivity, enabling unprecedented accuracy in peptide level measurements. This article provides a systematic overview of the underlying mechanisms, technical advantages, and optimization strategies of PRM in peptide quantification, and introduces the solutions developed by MtoZ Biolabs.
Fundamental Principles of Peptide Level Quantification Using PRM
Following enzymatic digestion, proteins are converted into a series of peptide fragments that serve as characteristic ions reflecting the abundance of their parent proteins. Quantification at the peptide level is accomplished by selectively detecting specific peptides via mass spectrometry and inferring protein abundance based on their chromatographic peak areas or intensities.
Key considerations include:
(1) Selecting representative (proteotypic) peptides that are unique, highly responsive, and stable.
(2) Ensuring consistent enzymatic digestion efficiency across samples while avoiding interference from post-translational modifications.
(3) Employing stable isotope–labeled peptides (SIL peptides) as internal standards to calibrate measurements and correct for sample and instrument variability.
Principles and Advantages of PRM in Peptide Quantification
PRM is a targeted quantification approach implemented on high-resolution mass spectrometers (e.g., Orbitrap). Its primary advantages include:
1. Comprehensive Fragment Ion Monitoring
Unlike SRM, which predefines 1–2 transitions, PRM simultaneously acquires all fragment ions, enhancing both specificity and quantitative flexibility.
2. High-Resolution Interference Reduction
Precise mass filtering (typically 0.005–0.01 Da) effectively suppresses background noise, ensuring accurate detection of low-abundance peptides.
3. Flexible Post-Acquisition Quantification
Researchers can retrospectively select optimal fragment ions based on data quality, improving quantitative robustness.
Key Determinants of Quantitative Accuracy
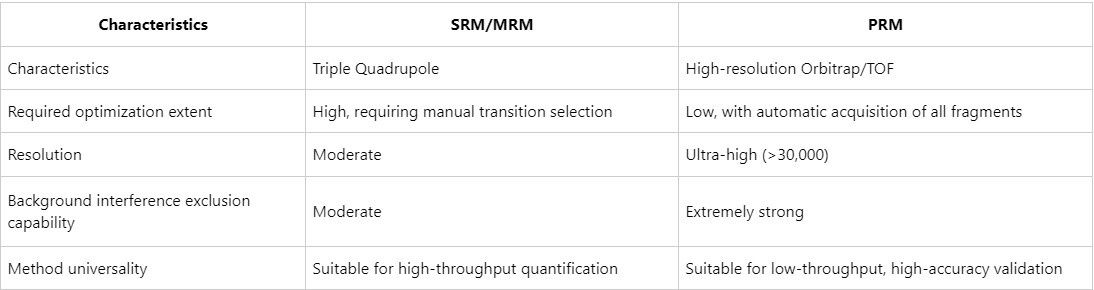
Comparative Advantages of PRM over SRM/MRM
MtoZ Biolabs has established a PRM-based targeted protein quantification platform built on the Orbitrap Exploris 480 system, featuring the following core capabilities:
With the advent of machine learning algorithms and AI-assisted methodologies, PRM data analysis is evolving toward higher automation and intelligence, enabling:
Furthermore, integrating medium-throughput automation platforms with curated peptide target databases will accelerate biomarker validation and provide robust evidence for precision medicine research. PRM technology has become a pivotal tool for precise peptide level quantification, demonstrating superior performance in studies requiring high reproducibility, low detection limits, and tolerance to complex biological matrices. MtoZ Biolabs continues to advance this field through professional mass spectrometry platforms and customized analytical services, empowering researchers to achieve greater accuracy and reproducibility in targeted protein quantification.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
How to order?







