Targeted Methylation Site Analysis Service
MtoZ Biolabs provides Targeted Methylation Site Analysis Service to accurately identify and characterize methylation at specific amino acid residues within defined proteins. This service focuses on the precise localization of methylation sites, determination of modification states, and evaluation of site occupancy levels. By combining optimized methylated peptide enrichment methods with high-resolution LC-MS/MS and expert bioinformatics analysis, MtoZ Biolabs helps researchers uncover site-specific regulatory mechanisms underlying protein activity, chromatin remodeling, and gene regulation.
Overview
Protein methylation is a critical post-translational modification that regulates gene expression, chromatin organization, and signal transduction. It primarily occurs on lysine (K) and arginine (R) residues, where methyl group addition alters protein structure and interaction patterns. Because each methylation site can exert unique biological effects, site-level analysis provides deeper insight into protein function than global methylation profiling.
Histone H3 methylation serves as a representative example of site-specific regulation. The lysine 4 residue (H3K4) can be mono-, di-, or tri-methylated, and each state corresponds to a distinct genomic function. H3K4me1 marks enhancer elements, H3K4me2 is associated with transcriptionally poised regions, and H3K4me3 is enriched near transcription start sites of active genes. Understanding these methylation site patterns reveals how individual residues contribute to transcriptional activation and chromatin dynamics.
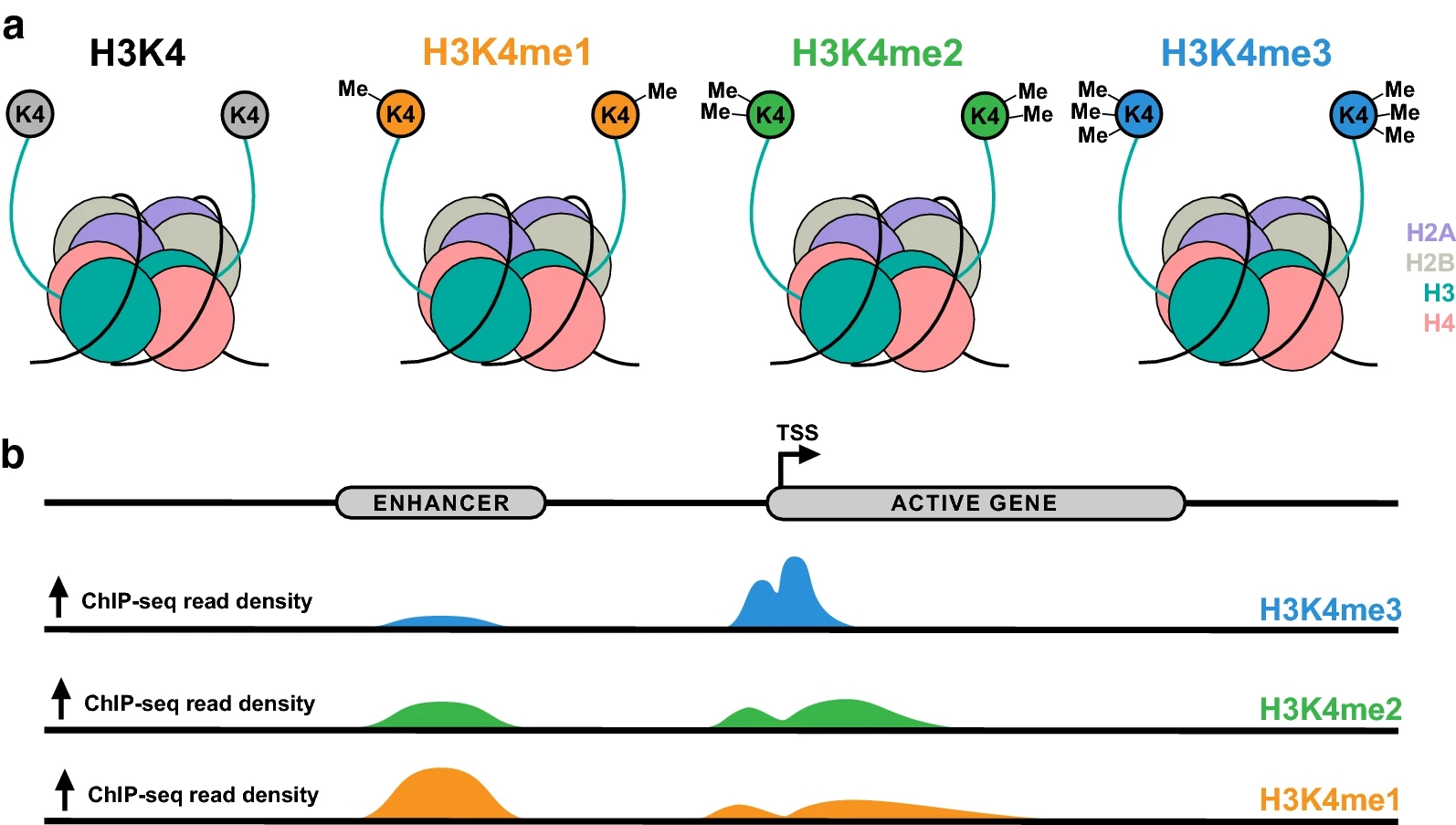
Collins, B. E. et al. Epigenet. Chromatin. 2019.
Figure 1. H3K4 Methylation Patterns and Their Genomic Distributions
Targeted Methylation Site Analysis Service at MtoZ Biolabs
MtoZ Biolabs offers a full range of analytical solutions dedicated to mapping and characterizing methylation sites on target proteins. Our platform integrates optimized sample preparation, efficient peptide enrichment, and precise LC-MS/MS detection to provide reliable site-level methylation data.
Our service includes:
✅ Precise Site Localization
Identification of methylation residues with amino-acid level accuracy using high-resolution MS/MS fragmentation patterns.
✅ Methylation State Characterization
Determination of mono-, di-, and tri-methylation at specific sites based on exact mass differences and spectral signatures.
✅ Occupancy Analysis
Evaluation of methylation site occupancy to estimate modification stoichiometry and detect partial or complete site methylation.
✅ Comprehensive Annotation and Reporting
Integration of methylation site information with protein domains, functional motifs, and biological relevance.
Workflow of Targeted Methylation Site Analysis Service
1. Sample Preparation
Proteins are denatured, reduced, alkylated, and digested into peptides under controlled conditions.
2. Methylated Peptide Enrichment
Targeted enrichment improves detection sensitivity and coverage. Strategies may include:
Immunoaffinity capture using anti-methyl lysine or anti-methyl arginine antibodies
Chemical derivatization to differentiate methylated residues
Chromatographic methods such as HILIC or SCX to enhance peptide separation
3. LC-MS/MS Analysis
High-resolution mass spectrometry platforms such as Orbitrap Fusion Lumos or Q Exactive HF are used for precise site localization.
4. Data Processing and Validation
MS/MS spectra are searched against curated databases to confirm methylation site identity and modification state.
5. Site Occupancy Evaluation
Site-specific occupancy is quantified to evaluate the relative abundance of modified and unmodified peptides.
6. Bioinformatics Annotation
Identified methylation sites are mapped to protein domains and pathways to reveal functional significance.
Why Choose MtoZ Biolabs?
· High-Confidence Site Localization
Our Orbitrap-based mass spectrometry systems provide accurate and reproducible identification of specific methylation sites with high localization confidence.
· Advanced Enrichment Strategy
Optimized enrichment strategies, including immunoaffinity purification, chromatographic separation, and chemical derivatization, ensure efficient recovery of low-abundance methylated peptides.
· Comprehensive Bioinformatics Interpretation
Each project includes in-depth annotation of methylation sites, integrated with protein structure, domain information, and biological pathways for meaningful functional insights.
· Customized Project Design
Flexible experimental design tailored to your target protein, sample type, and research goals to ensure optimal analytical performance.
· Experienced Scientific Support
Our expert proteomics team provides end-to-end technical consultation, data validation, and interpretation throughout the project lifecycle.
Applications of Targeted Methylation Site Analysis Service
Targeted methylation site analysis enables detailed investigation of protein regulation and epigenetic control across multiple research fields:
Epigenetics: Mapping histone methylation sites for chromatin and transcriptional studies.
Transcriptional Regulation: Identifying methylation sites that influence transcription factor binding.
Signal Transduction: Investigating site-specific methylation in signaling enzymes or regulators.
Oncology: Detecting abnormal methylation sites involved in cancer progression.
Drug Discovery: Supporting the development of methylation-targeted therapeutics.
Start Your Project with MtoZ Biolabs
MtoZ Biolabs is committed to delivering accurate and biologically meaningful methylation site data to advance your research. Whether you are exploring histone methylation patterns or studying site-specific regulation in non-histone proteins, our analytical expertise and state-of-the-art instrumentation ensure dependable results.
Contact us today to discuss your project or request a customized quotation. Our team will assist you in developing a targeted workflow for precise methylation site identification and analysis, empowering your research with high-resolution proteomic insights.
FAQ
Q1: What types of samples are suitable?
MtoZ Biolabs accepts a wide variety of samples for Targeted Methylation Site Analysis, including cell lysates, tissue extracts, recombinant proteins, and synthetic peptides. Both native and modified proteins can be analyzed to identify methylation sites on specific targets. Recommended sample amounts are ≥500 µg of total protein for lysates or ≥50 µg of purified protein.
Q2: What is the service general workflow?
Q3: How should I prepare my samples?
Q4: What data formats are provided?
Comprehensive deliverables are provided to support downstream analysis, including:
Raw LC-MS/MS data files
Processed identification and site mapping tables (Excel or CSV format)
Annotated spectra and peptide summary reports (PDF format)
Customized data interpretation and visualization according to project needs
Related Services
How to order?







