SUMOylation proteomics Analysis Service
SUMOylation proteomics Analysis Service accurately identifies SUMOylation modification sites and target proteins, analyzes the dynamic regulatory mechanism of SUMOylation in specific biological processes, and provides important clues and new targets for disease research and drug development. SUMOylation is an essential type of post-translational modification (PTM), characterized by the covalent attachment of SUMO proteins to lysine residues on target proteins. This modification regulates protein stability, activity, subcellular localization, and interactions with other molecules. SUMOylation plays critical roles in various biological processes, including cell cycle control, gene expression regulation, protein stability maintenance, and nucleocytoplasmic transport. It is also closely associated with numerous diseases, such as cancer, neurodegenerative diseases, and infectious diseases. Utilizing specific SUMOylation enrichment techniques combined with high-resolution mass spectrometry, MtoZ Biolabs provides SUMOylation Proteomics Analysis Service to accurately identify and quantify SUMOylated proteins, helping researchers uncover the dynamic changes and regulatory mechanisms of SUMO modification in specific biological processes.
Analysis Workflow
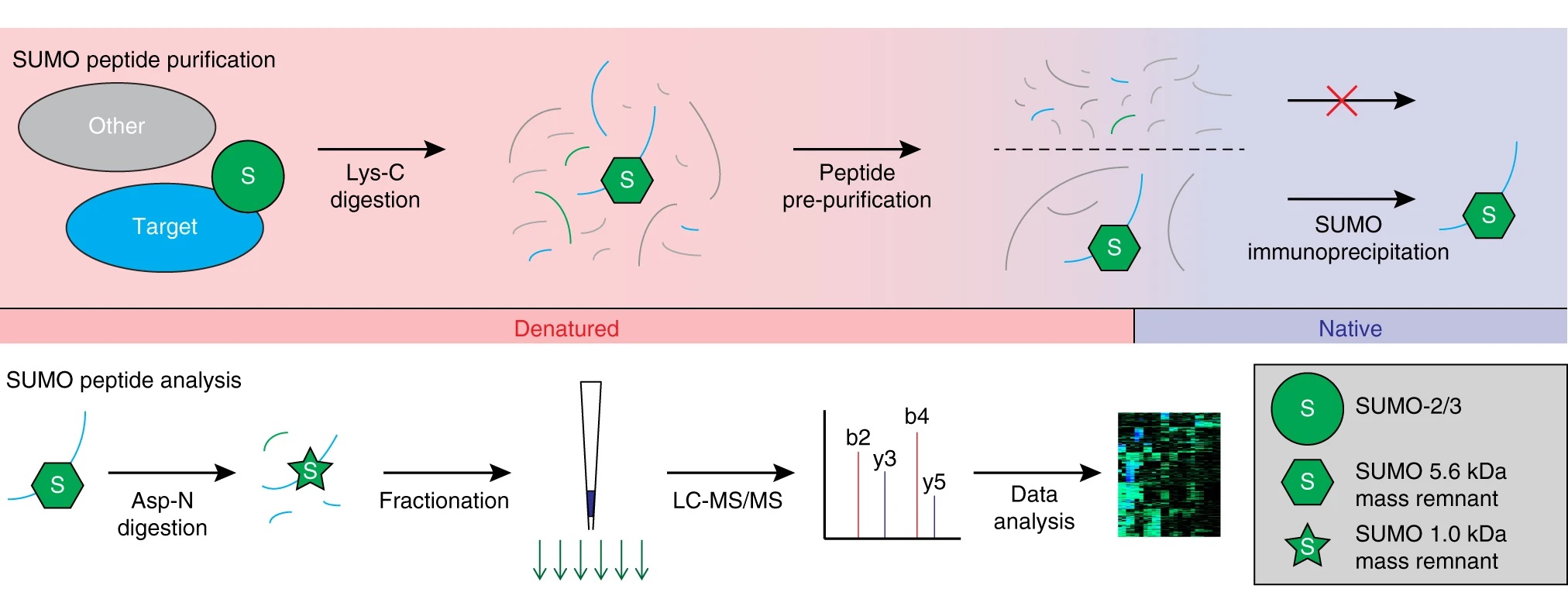
Hendriks I A. et al. Nature Communications. 2018.
1. Sample Preparation
Extract proteins and remove non-specifically bound proteins to ensure the integrity of total protein and SUMOylation modifications. Digest proteins into peptides suitable for mass spectrometry analysis.
2. Enrichment of SUMOylated Proteins
Employ anti-SUMO antibodies or SUMO affinity resin to specifically enrich SUMOylated proteins.
3. LC-MS/MS Analysis
Separate the complex peptide mixtures using nano-LC. Analyze peptides using high-resolution mass spectrometry to obtain detailed mass spectra.
4. Data Processing and Analysis
Identify SUMOylated peptides through database searches and advanced data analysis tools, precisely localizing modification sites and determining modification states.
Perform quantitative analysis to study dynamic changes of SUMOylation under various conditions.
Utilize enrichment analyses, functional annotation, and network analysis to elucidate the roles of SUMOylated target proteins in signaling pathways and biological functions.
Services at MtoZ Biolabs
MtoZ Biolabs, an integrated Chromatography and Mass Spectrometry (MS) Services Provider, provides advanced proteomics, metabolomics, and biopharmaceutical analysis services to researchers in biochemistry, biotechnology, and biopharmaceutical fields. Our ultimate aim is to provide more rapid, high-throughput, and cost-effective analysis, with exceptional data quality and minimal sample consumption. Leveraging advanced mass spectrometry and rigorous data analysis methods, MtoZ Biolabs is dedicated to providing clients with a high-quality SUMOylation Proteomics Analysis Service helping comprehensively analyze the dynamic changes and biological significance of SUMOylation modifications. Our service empowers fundamental research and clinical translation, providing robust support for scientific discovery and drug development.
Applications
1. Disease Mechanism Research
Revealing the role of SUMOylation modifications in cancer progression, such as regulating the activity of tumor suppressors or oncogenes.
2. Signal Transduction and Gene Expression Regulation
SUMOylation Proteomics Analysis Service can be used to investigate dynamic SUMOylation changes in cellular signaling pathways and their impacts on processes like cell cycle control and DNA repair.
3. Discovery of Novel Drug Targets
Identifying critical SUMOylated target proteins, thereby uncovering potential drug targets.
4. Development of Molecular Diagnostic Biomarkers
SUMOylation Proteomics Analysis Service can analyze changes in SUMOylation modification patterns, identifying disease-related diagnostic and prognostic biomarkers.
FAQ
Q. How can the enrichment efficiency of SUMOylated proteins be improved?
Due to the low abundance and dynamic nature of SUMOylation modifications, enrichment is a critical step. The following approaches can significantly enhance enrichment efficiency:
1. Use of Highly Specific Anti-SUMO Antibodies or Affinity Resin: Select reagents that exhibit high affinity for target SUMO isoforms and low background binding.
2. Optimization of Sample Preparation: Apply specific conditions to stabilize SUMOylation modifications prior to cell lysis to minimize modification loss.
3. Improvement of Protein Recovery: Incorporate multiple rounds of affinity purification or combine antibodies targeting multiple SUMO isoforms to increase the total amount of enriched proteins.
Q. How can the authenticity of SUMOylation sites be validated?
After identifying SUMOylation sites, their authenticity can be validated through the following methods:
1. Mutational Analysis: Conduct site-directed mutagenesis to replace lysine residues with non-modifiable amino acids (e.g., arginine) and observe whether the SUMOylation signal is abolished.
2. Independent Enrichment Strategies: Validate the same modification sites using different affinity methods (such as comparing anti-SUMO antibodies with SUMO-binding domain tags).
3. Functional Experiments: Assess whether SUMOylation at identified sites affects the protein’s subcellular localization, stability, or function, indirectly confirming modification authenticity.
4. Repeated Detection: Identify the same modification sites across multiple experimental replicates or conditions to ensure they are not incidental background signals.
By implementing these validation steps, the accuracy of SUMOylation sites can be effectively confirmed, providing a robust foundation for subsequent functional analyses.
Deliverables
1. Comprehensive Experimental Details
2. Materials, Instruments, and Methods
3. Total Ion Chromatogram & Quality Control Assessment (project-dependent)
4. Data Analysis, Preprocessing, and Estimation (project-dependent)
5. Bioinformatics Analysis
6. Raw Data Files
Case Study
This study utilized SUMOylation proteomics analysis to identify SUMO-modified proteins in the Drosophila ovary. The results revealed that SUMOylation modifications were broadly present in proteins involved in heterochromatin regulation and the piRNA pathway. Further analyses indicated that SUMOylation of several piRNA pathway proteins was dependent on the presence of Piwi protein. This study highlights the crucial role of SUMOylation modifications in epigenetic regulation and genome defense, providing new insights into the function of SUMOylation in heterochromatin formation and transposon silencing.
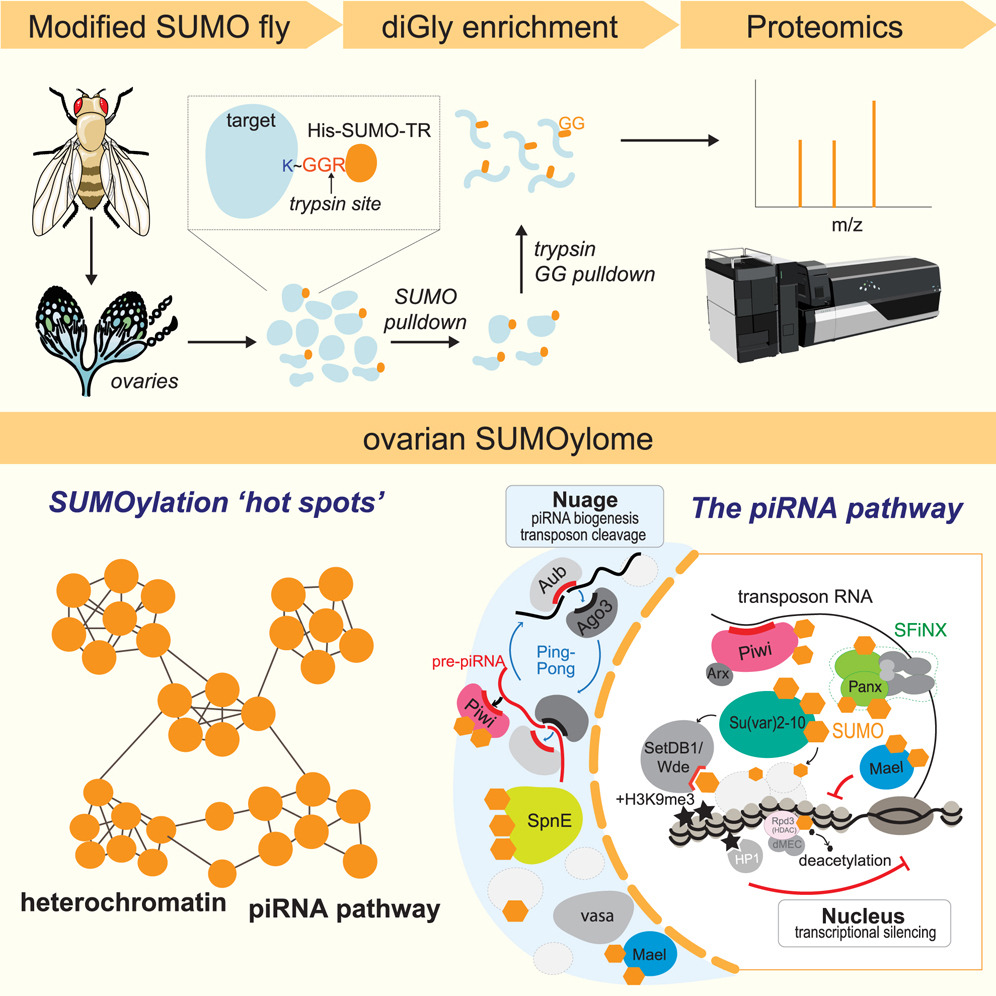
Ninova M. et al. Cell Genomics. 2023.
MtoZ Biolabs, an integrated chromatography and mass spectrometry (MS) services provider.
Related Services
Quantitative Phosphoproteomics Service
How to order?







